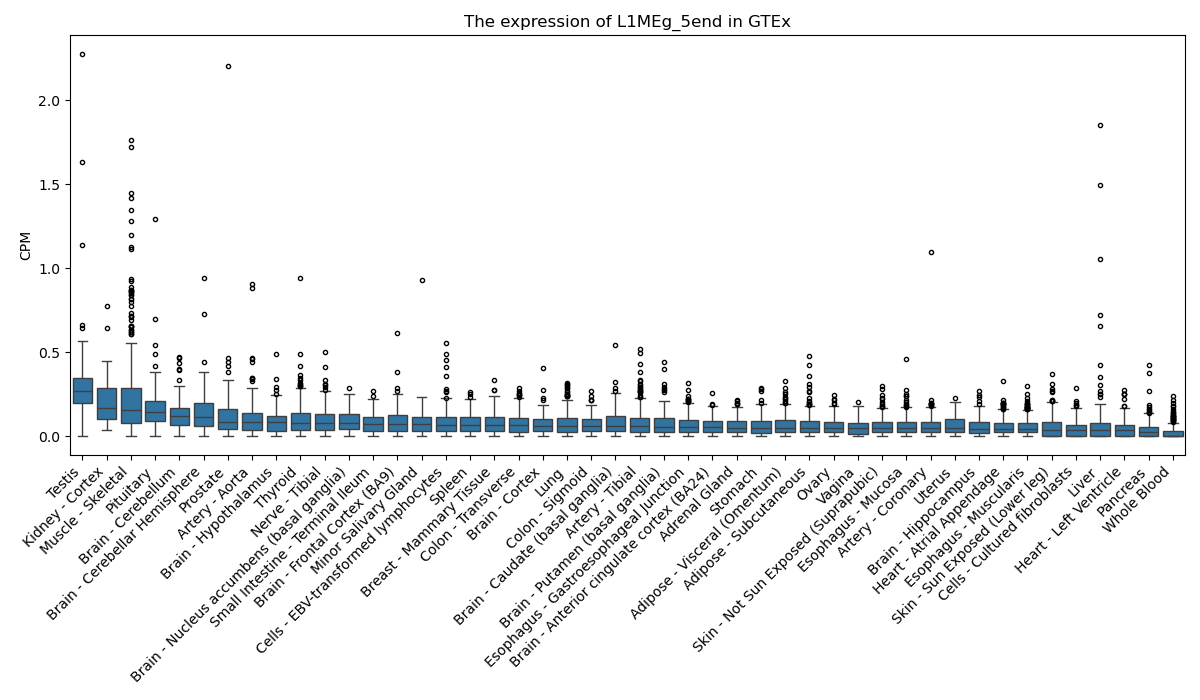
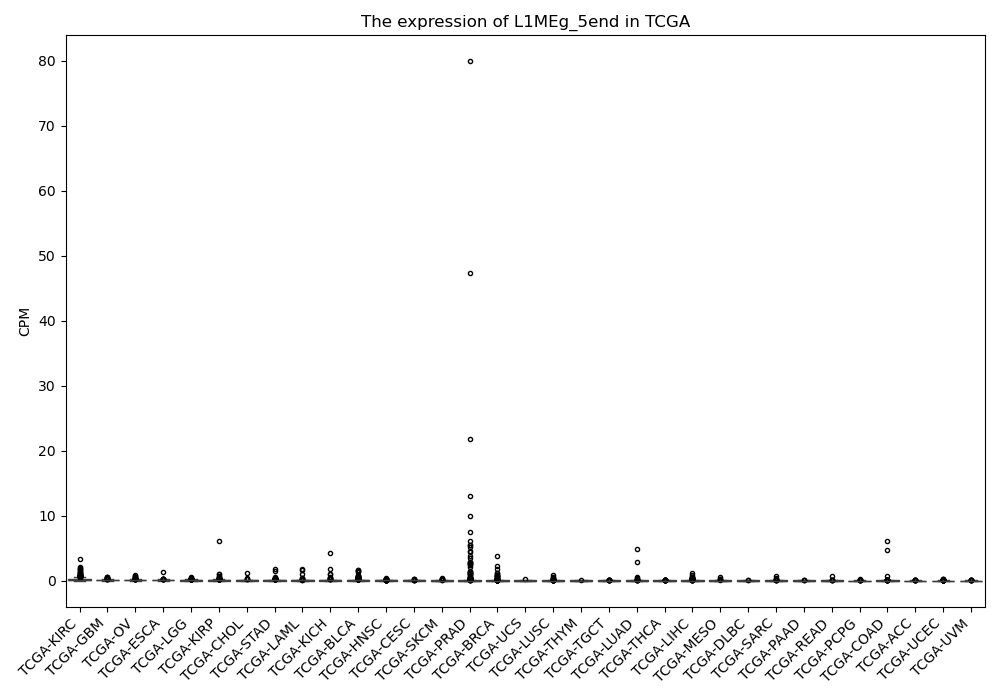
L1MEg_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000311 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Theria_mammals |
| Length | 2101 |
| Kimura value | 29.58 |
| Tau index | 0.7568 |
| Description | 5' end of L1 retrotransposon, L1MEg_5end subfamily |
| Comment | - |
| Sequence |
TTCCGCTTCCGGCAATGGCGGACTAGNTTGTTCAGACCAACCCTCCCGCTGAGAACAACTAGAAAAGCTGGACAAAATATAAAAAACATCTGTTTGAAGGCATCGGAGAGCTACCAAGGCAGCGAGGAATTTGAGGGGCCAAGATCCCGGAGAGAAGGGAAGCCCAGAGAGGTGAGCCCGACATTCGGNGCCGCTTTTCCCCTCGAGGCATTTGCCGATTCCGAAAGCGGCGGCTGAGAGGCTGAGAAGCTGAGCAGAGCTTTCGGCAGTCTCACGGGGCTGGGGAGACAAAAATTGGAGTTCAGGGCCCGCCAAGGAGGAGGGGCCCTGGTAAACCCCAGCTTTCAGTTGGGACCCCGAAGGGCTACACCCTAGGAGTAAGGGCGAACCGGAAATAGACCAGCCCTCACAAAGACTGAAGCCCAGCTTCGAATCANCTCAATCCCTGATTGGATTAAGGTGATCTGATTGCTAGTGCCCCTAGCTGCCTGCCAGAAGCAAAAGTAAATCCTCTCTGGAGGAAGATAACATCATCCAGAGCCTCAAATTATCTCTACAATTTTTCATATACAATGTCCGGCATTCAATCAAAAATNACCAGGCATACNAGGAGACAAGACAAATGACCGAAAANCAAGAGAAAAACAGACAATAGAAACAGACCCACAGGNGATCCAGATATTGGAGTTATCAGACACGGACTTTAAAATAACTATGATTAATATGTTCAAGAAAATAAANGACAAGATGGAGAATTTCAGCAGAGAACTGGAANCTATAAAAAAGAATCAAATGGAAATTCTAGAACTGAAAAATATACAATAACTGAAATTAAGAACTCAATAGATGGGTTTAACAGCAGATTAGACACAGCNGAAGAGAGGATTAGTGAACTGGAAGATAGGTCAGNAGAAAATATCCAGACTGAAGCACAGAGAGANAAAAAAATGGAAAATACAGAAAAGAGCGTAAGAGACATATGGGACACGGTGAAAAGGTCTAACATACGTGTAATTGGAGTCCCAGAAGGAGAGGAGAGAGAGAATGGGGCAGAAGCAATATTTGAAGAGATAATGGCCGAGAATTTTCCAAAACTGATGAAAGACATCAANCCACAGATTCAAGAAGCTCTACGAACCCCAAGCAGGATAAATACAAAGAAAACCACACCTAGGCACATCATAGTAAAACTGCTGAAAACCAAAGACAAAGAGAAAATCTTAAAAGCAGCCAGAGAAAAAAGACACATTACCTTCAAAGGAGCAACAATAAGACTGACAGCTGACTTCTCAACAGAAACAATGGAAGCCAGAAGACAATGGAATGACATCTTTAAAGTGCTGAAAGAAAATAACTGCCAACCTAGAATTCTATACCCAGGAAAATATCCTTCAAAAATGAAGGCGAAATAAAGACATTTTCAGACAAACAAAAACTGAGAGAATTCGTCACCAGCAGACCTGCACTAAAAGAAATACTAAAGGAAGTTCTTCAGGCAGAAGGAAAATGATCCCAGATGGAAGCACGGAAATGCAGGAAGGAATGAAGAGCAACGGAAAGGGTAAATATGTGGGTAAATCTAAATGAATATTGACTGTATAAAACAATAATAATAATGTCTTGTGGGGTTTAAAATATATATAGAATTAAAATACATGACAACAATAGCACAAAAGGGAAGAGAGGGTAAATGGAGTTAAAGTGTTCTAAGGTCCTTGCATTGTCCGGGAAGTGGTAAAGTACTAATTTATATTAGACTTTAATAAGTCAAGGATGCATGTTGTAATCTCTAGGGTAACCACTAAAAGAATAGTAAAAGAATGTATAACTAACAAGCTAATAGAGAGGGAAAATGGAATAATAAAAATATTCGATTAATCCAAAAGAAGGCAAGAAAAAAGGAAAAAANAGAAAGAAAAACAGATGGGACAAATAGAAAACAAATAGTAAGATGGTAGATTTAAACCCAAATATATCAGTAATTACATTAAATGTAAATGGACTAAATACTCCAATTNAAAGANAAAGATTGTCAGACTGGATTTAAAAACAAAACCCAACTATATGCTGCTTACAAGAGACACACCTTAAATATAAGGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MEg_5end | ZNF85 | 1779 | 1790 | - | 15.97 | GAGATTACAACA |
| L1MEg_5end | PI | 1893 | 1906 | + | 15.89 | AGAAAAAAGGAAAA |
| L1MEg_5end | CDF5 | 1885 | 1905 | - | 15.89 | TTTCCTTTTTTCTTGCCTTCT |
| L1MEg_5end | TFAP2B | 198 | 210 | + | 15.87 | TCCCCTCGAGGCA |
| L1MEg_5end | DOF1.5 | 1891 | 1906 | + | 15.86 | CAAGAAAAAAGGAAAA |
| L1MEg_5end | sage | 83 | 93 | + | 15.79 | AAAACATCTGT |
| L1MEg_5end | TAL1::TCF3 | 85 | 94 | + | 15.75 | AACATCTGTT |
| L1MEg_5end | ZNF148 | 316 | 325 | - | 15.58 | CCCCTCCTCC |
| L1MEg_5end | pnt | 388 | 396 | + | 15.57 | ACCGGAAAT |
| L1MEg_5end | TFAP2C | 198 | 210 | + | 15.50 | TCCCCTCGAGGCA |
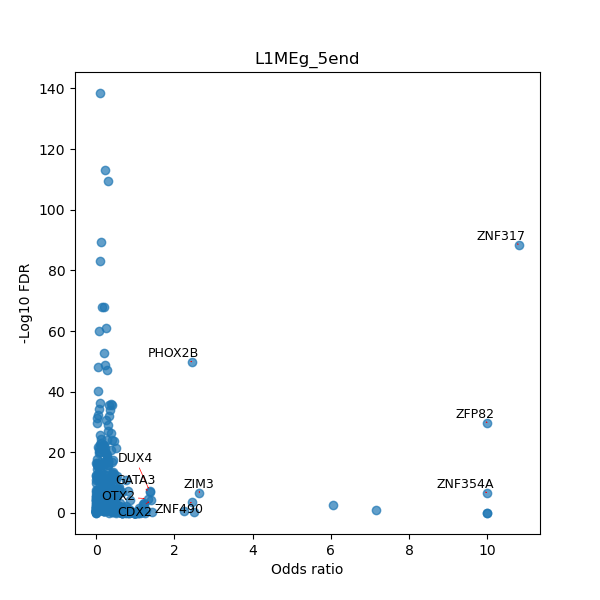
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.