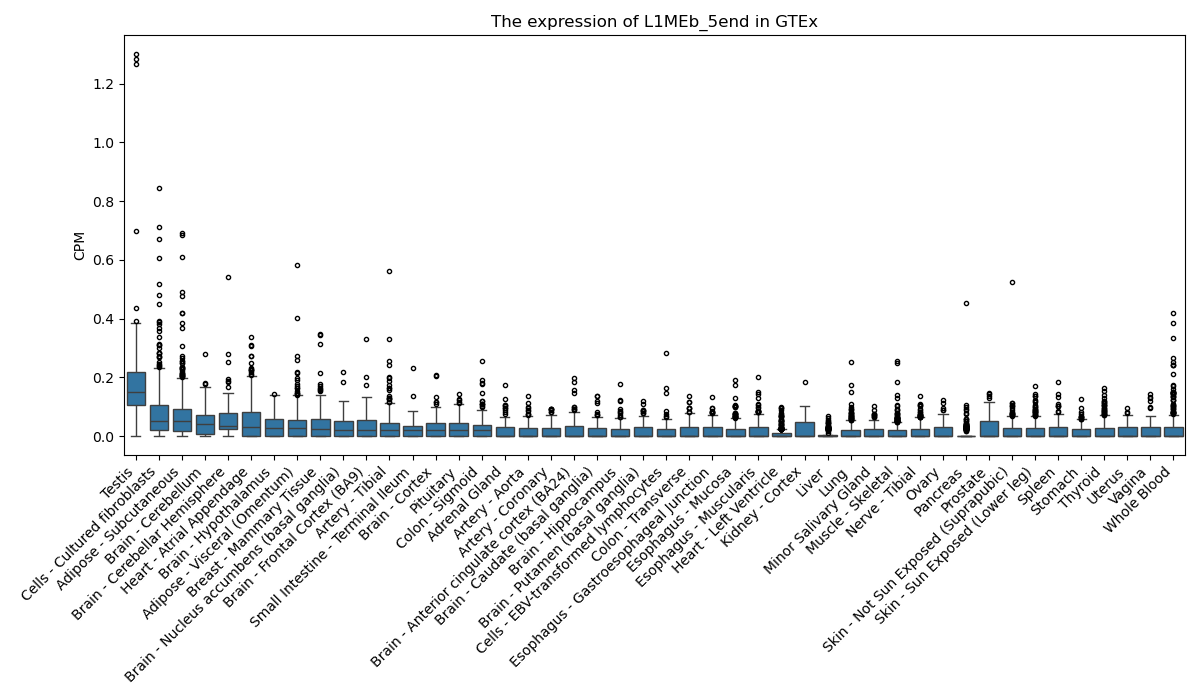
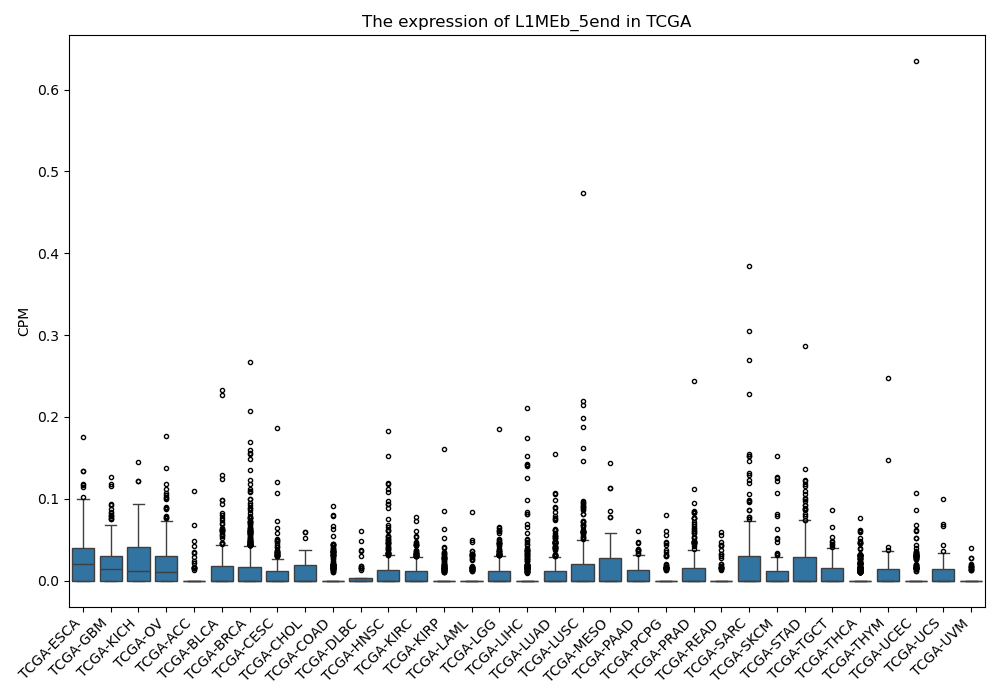
L1MEb_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000774 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 1089 |
| Kimura value | 28.63 |
| Tau index | 0.9360 |
| Description | Partial 5' end of L1 retrotransposon, L1MEb_5end subfamily |
| Comment | 5' end of L1ME1 or L1ME2 LINE1. Average divergence from consensus is 26%. L1MEB_5 is 70% similar to L1MDA_5 sequence. |
| Sequence |
CAGANTTCCGCTTCCGGTAATGGCGGAGTAAGCTCCTATCGGACCAACCCTCCCGCAGATAACAACTATAAACTCTGGACAAAATACAAAAAACAACTACCTGAAGGCACTGGAGAGCGACCAAAAGCAGGCAGANACTGGAGGGGAGTCGACACTTGGAAGAAGGGAACGGCACNGGGTGAGTTCCCATTTTTACGGCTTTTNGCCTGAGGGCAGGCCNCAGTCGGTGCGNCGTACAGGTGGCTAAAACTCCAGTAGAAAACCCGCAGTCTTNCTGGCTTGAAGAACCAGAGGACAGAGTTCGGGGCAACCACAGCCGCTGGAAAGTGAGGGGGAAATCCCGGAAAGGAGAGAGCCAGAGAGAGGAGCCCCAAATTCTGCGTATAAACTCTGCCCAAATCTCTGGCTGACCCCTGAACCACGCATGCGCGGGGCAGACTCCAAGCAGCCCAGCTAAAGATAAAAGAACTGAACTGAGATTTNAGCTGCCGCCCAAGAGACAGAGTTTGNAGTTTGAGTCCAGCCAAGTTAACTGCCTGCTAAAACAAAANAANCAACACTCTTCAGAGGAANATAACAGAATCCAGAGTCTCTACAACGTANCATTCACAATGTCCAGGATACAATCCNAAAATTATNCNAGATANGAAGAAACAGGAAAAATGTGACCCATNCTCAAGAGAAAAGAAAATAGAAACCGACCCCGAGATGACCCAGATGTTGGAATTAGCAGACAAGGACTTTAAAGCAGCTATTATAAATATGTTCAANGANNTAAAGGAAAATATGCTCATAATGAATGAAAAGATAGGAAATCTCAGCAGAGAAATAGAAACTATAAAAAGAACCAAATGGAAATTCTAGAACTGAAAAATACAATATCTGAAATAAAAAATTCACTGGATGGGCTTAACAGCAGANTGGAGATGACAGAGGAAAGAGTCAGTGAACTTGAAGATAGATCAATAGAAATTATCCAATCTGAAGAACAGAGAGAAAAAANATTTAAAAAAAAAGAGCAGAGCCTCAGNGACCTGTGGGACAATATCAAAAGGTCTAACATACGTGTAATTGGAGTCCCAGAAGGAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MEb_5end | RARA::RXRG | 699 | 715 | - | 18.79 | GGGTCATCTCGGGGTCG |
| L1MEb_5end | ZNF530 | 344 | 357 | + | 17.82 | GAAAGGAGAGAGCC |
| L1MEb_5end | Dif | 333 | 342 | - | 17.39 | GGGATTTCCC |
| L1MEb_5end | Dif | 333 | 342 | + | 17.21 | GGGAAATCCC |
| L1MEb_5end | ZFP14 | 103 | 117 | + | 17.15 | GAAGGCACTGGAGAG |
| L1MEb_5end | Nrf1 | 421 | 432 | - | 16.76 | CCGCGCATGCGT |
| L1MEb_5end | ewg | 421 | 430 | + | 16.43 | ACGCATGCGC |
| L1MEb_5end | ewg | 421 | 430 | - | 16.43 | GCGCATGCGT |
| L1MEb_5end | STAT1::STAT2 | 825 | 837 | - | 16.20 | AGTTTCTATTTCT |
| L1MEb_5end | ETS2 | 10 | 18 | - | 16.00 | ACCGGAAGC |
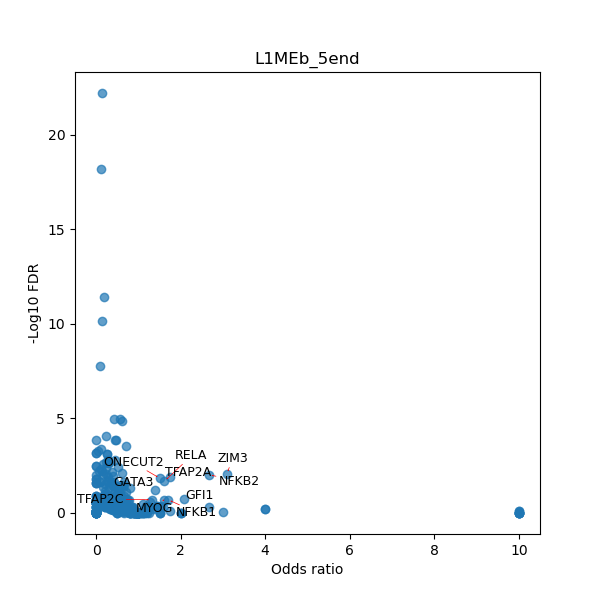
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.