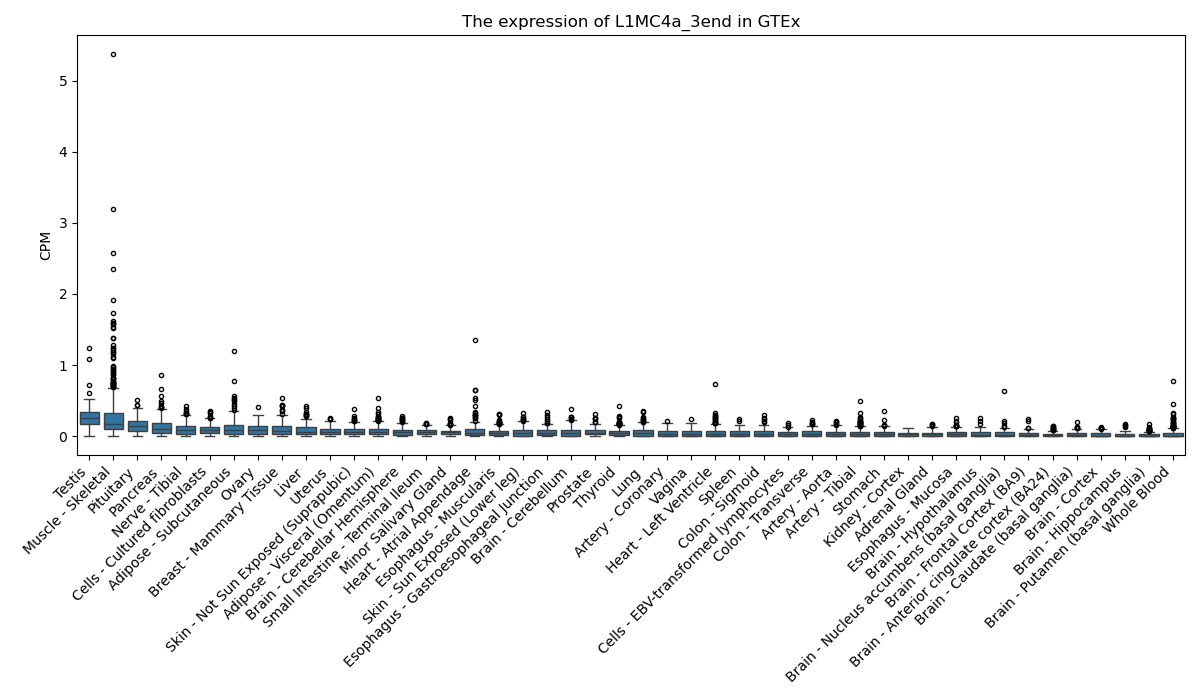
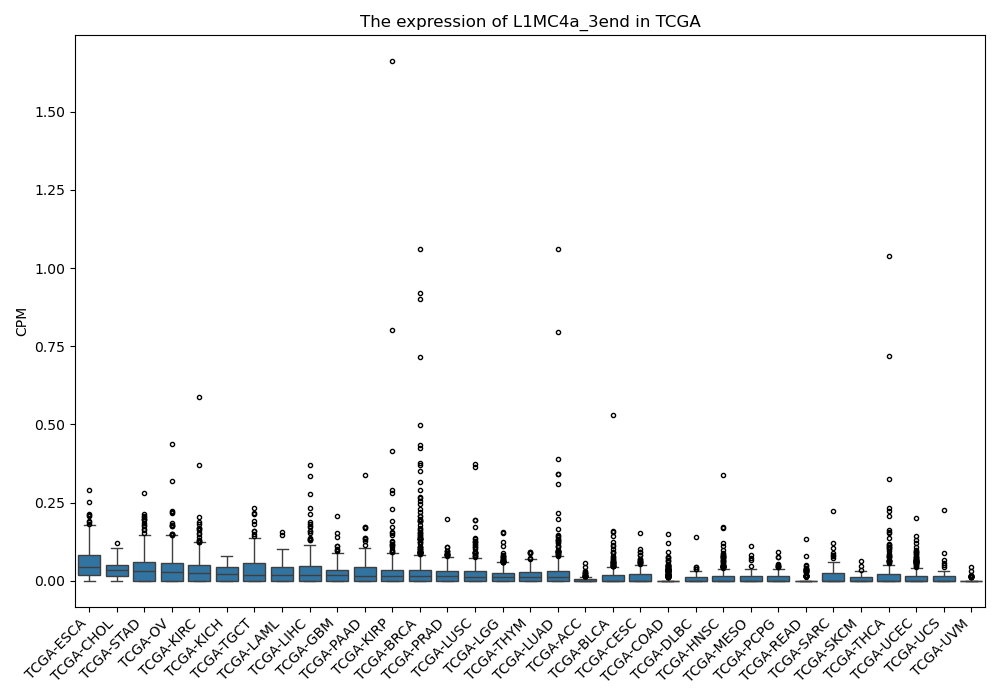
L1MC4a_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000279 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2629 |
| Kimura value | 23.95 |
| Tau index | 0.8175 |
| Description | 3' end of L1 retrotransposon, L1MC4a_3end subfamily |
| Comment | - |
| Sequence |
CTAATATCCCTAATATATAAAGAACTCTTAAAANTNGAAAGNAAAAGACCAAAAACCCGATAGAAAAATGGGCAAAAGACATGAACAGACAATTCACAAAAAGANATANAAATGGCCCTTAAACATATGAAAAGATGTTCAACCTCACTCATAATAAGAGAAATGCAAATTAAAACTACACTGAGATACCATTTCTCACCTATCAGATTGGCAAAAATTCAAAAGTTTGACAACACACTCTGTTGGCGAGGCTGTGGGGAAACAGGCACTCTCATACATTGCTGGTGGGAATGCAAANTGGTACAACCCTTNTGGAGGGGAATTTGGCAATATCTAACAAAACTACATATGCATTTACCCTTTGACCCAGCAATCCCACTTCTAGGAATTTACCCTGAAGATACACCTCCAACATGTACAAAATNACATATGCACAAGGTTATTCATTGCAGCATTGTTTGTAATTGCAAAATATTGGAAACAACCTAAATGCCCATNNATAGGAGANTGGTTGAATAAACTATGGTACATCCACACAATGGAGTACTATGCAGCTGTAAAAAAGAATGAGGAAGATCTCTATGAACTGATATGGAGTGATTTCCAGGATATATTGTTAAGTGAAAAAAGCAAAGTGCAAAAGAGTATNTATAGTATGCTACCTTTCGTGTAAGAAAGAAGGGAAAATAAGAAAATATACATGTATCTGCTCATTTGTGCAAAAAGAAACACAGGAAGGATAAACCAGAAACTAATGAGATTGGTTACCTACAGGGGGTGGGTGGGAATGGGGAGAAAGAATGGGGGATGGGAACGGGATGGNAGGGATGAGGAGGGAGTGACACTTCTCTGAGTATACCTTTTTGTATAGTTCTGACTTTTAGAACCATGNTAATGTTTCACATACCTCAAAAATAAATAATTAAAATCAACCAGGATGTGGGGGGAACCCAAAATGGAATACAAACAGNAACAAATGAACCTAACTGTATTNCAAATGAATAACATAACCACACTGAAGGGGATGGGGAAGAAAAGAACTAACCTAAGTAACTTTGGAAAACAGTATTTTGACTGNATACTGTAAGGCTAAAGACAAAAAGAACTGTACACAAATACTGTACTCTAGTTAGTAAATTTGTTTCTCACAGGGGTATGGGTTAGCAATTCTGAAACTACTTTATGTGTATNCTAGGATTGAACAAATAAGTAAATATATTGTAGATAATGAGAGCCAGGTTTCTCACTGTCGGAGAAAGAAGTTACAAATAAGGAAAGGGGGAAGGCTAGAATGAACCCTGTGGTGTTGGATTGGAATCGGAGGTATCAGTATGAACTCATGGTTTTTAACATATATACAGATAGATAGATACAGAAATAAGATATAGATGTATGTGTATGCATGGGTTAGTATACATACATATATTTCCTAGCTCTGTCCGCTGAGAGGGCCTAGAAGCAATGACACCCCAGTAGCAATGAGCACACCTAGCGCCCAGATCTTGGTTTCTAAATACCATTCTCCAATAAAAGGAACCAGGGCTCCTTGGAGAAATGGCTGATTCCAGGGCTGGGGCAGGGAAAATACAAGATGAGCCTGGAACATCTTGTGGTGCCAGAAAGTAAGGAAGTGCTCAAAAAANGATGGGGGCATGTCAAAAGGACACAGGAGCCAACCTGAAGGAGCTCCCAATGGCCAAANCTGGAACAATTTGAGCAACAAAATAAATAATGATAGTANTGGATTATAACCCATAGAATAAAATAAATATCCATGAGTCCATACTGATATAAATAAATAATTGAATAAATAAATAAATGGGGGAGAAGGGACAGCTCTTCCTTACAGNAGAATTCCAATTAATAAATGTAGAAGGAATGANGGAAATAGAAAATCACCATTAGGCAAACACCACAGTAATAATTGTTGCAGGCAAGATCCACCGATGGATGCTAAAATTAGTGGGCGAAAGTTTGAGGAGAAACAGGATATTTGCATAGTCTCAAAGTATCTCCCCCAAGATATTTATTAATTACAAAGGGAAAAATAGTAACTTTACAGTGGAGAAACCCGGCAGACACCACCTTAACCAAGTGATCAAGGTTAACATCACCAGTAATAAGACATATCGACATCATGTACCCCCTGATATGATGCACTGAGAAGGGCACAACATCACTTCTGTGGTATTCTTGCCAAAAATGCATAACCTCAATCTAATCATGAGAAAACATCAGACAAACCCAAATTGAGGGACATTCTACAAAATAACTGACCAGTACTCTTCAAAAGTGTCAAGGTCATGAAAGACAAGGAAAGACTGAGGAACTGTCACAGATTGGAGGAGACTAAGGAGACATGACAACTAAATGCAATGTGGGATCCTGGATTGGATCCTGGAACAGAAAAAAGGACATTAGTGGAAAAACTGGTGAAATTCGAATAAGGTCTGTAGTTTAGTTAATAGTATTGTACCAATGTTAATTTCCTGGTTTTGATAATTGTACTATGGTTATGTAAGATGTTAACATTAGGGGAAGCTGGGTGAAGGGTATACGGGAACTCTCTGTACTATTTTTGCAACTTTTCTGTAAGTCTAAAATTATTTCAAAATAAAAAGTTAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MC4a_3end | POU2F3 | 1992 | 2000 | - | 15.87 | TATGCAAAT |
| L1MC4a_3end | Esrrg | 2296 | 2304 | + | 15.85 | TCAAGGTCA |
| L1MC4a_3end | SPL8A | 542 | 550 | - | 15.77 | ATAGTACTC |
| L1MC4a_3end | Su(H) | 1141 | 1150 | - | 15.71 | TGTGAGAAAC |
| L1MC4a_3end | SOC1 | 1265 | 1277 | - | 15.66 | TTTCCTTATTTGT |
| L1MC4a_3end | PI | 1265 | 1278 | + | 15.62 | ACAAATAAGGAAAG |
| L1MC4a_3end | HOXB13 | 1524 | 1532 | + | 15.62 | CCAATAAAA |
| L1MC4a_3end | POU3F4 | 1992 | 2000 | - | 15.57 | TATGCAAAT |
| L1MC4a_3end | ZBTB32 | 1116 | 1125 | - | 15.47 | AGTACAGTAT |
| L1MC4a_3end | SGR5 | 1089 | 1101 | + | 15.39 | GCTAAAGACAAAA |
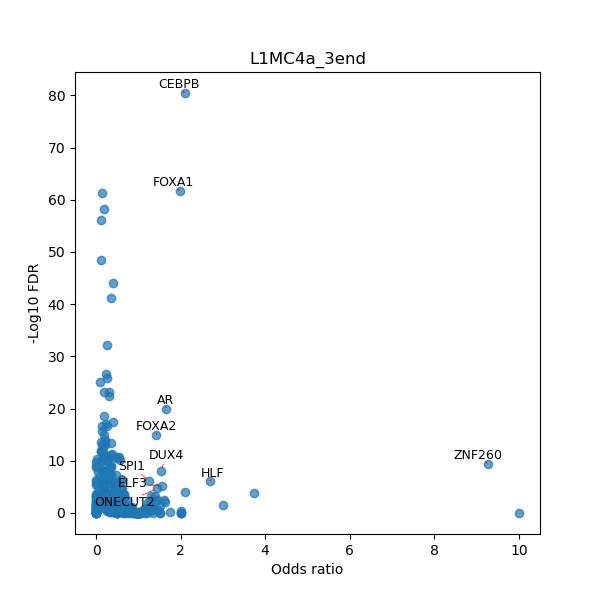
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.