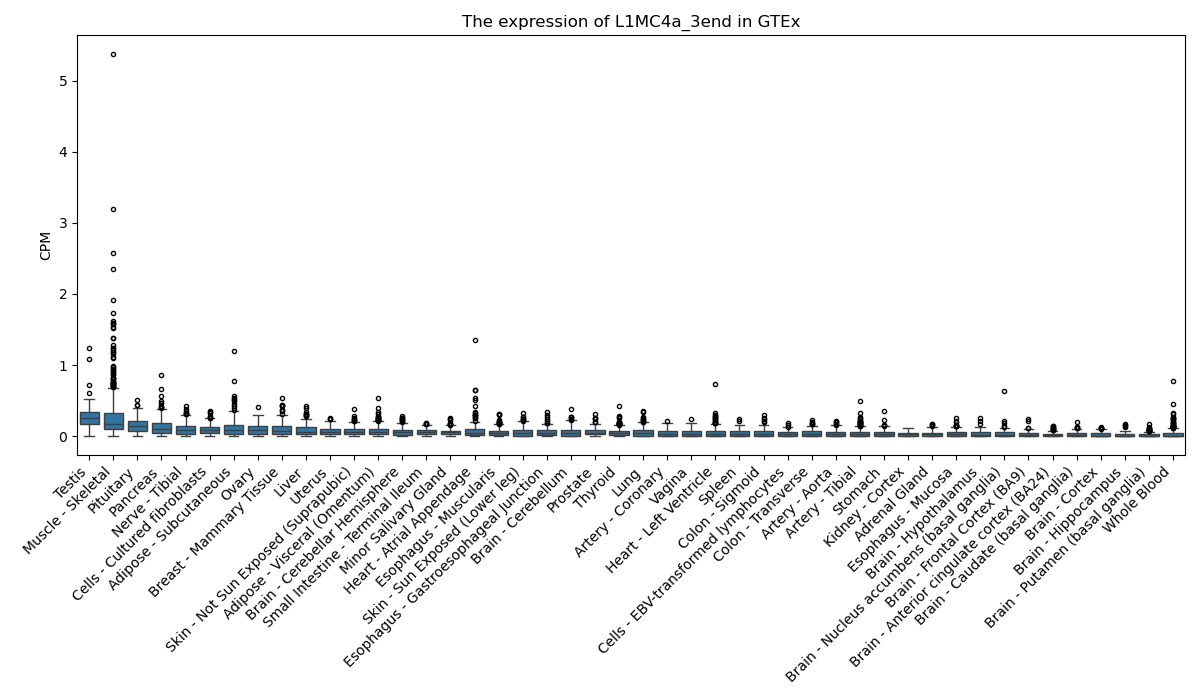
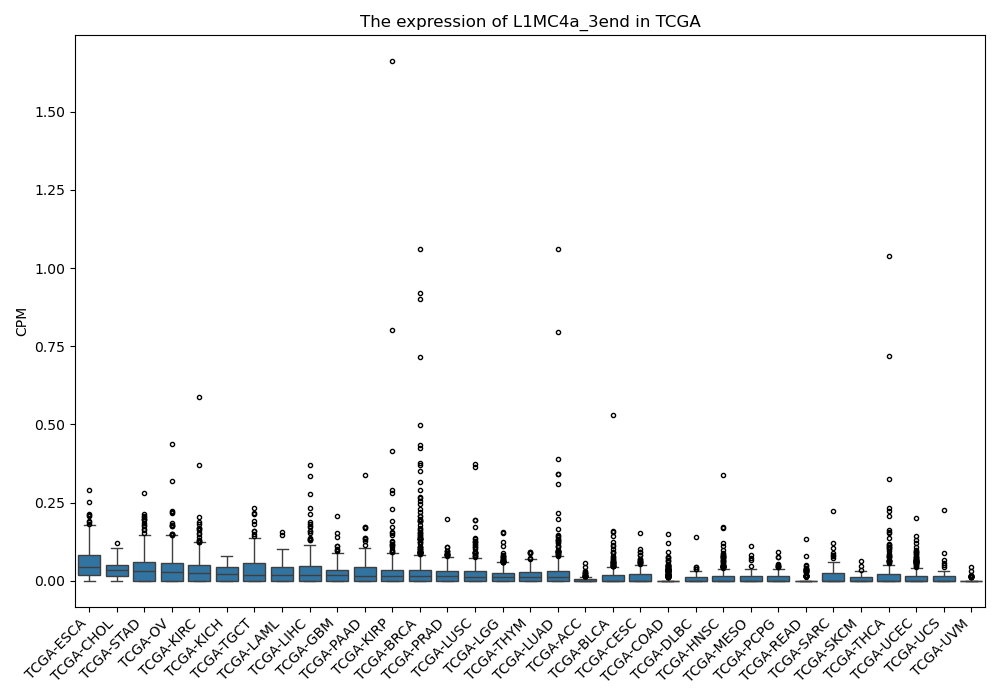
L1MC4a_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000279 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2629 |
| Kimura value | 23.95 |
| Tau index | 0.8175 |
| Description | 3' end of L1 retrotransposon, L1MC4a_3end subfamily |
| Comment | - |
| Sequence |
CTAATATCCCTAATATATAAAGAACTCTTAAAANTNGAAAGNAAAAGACCAAAAACCCGATAGAAAAATGGGCAAAAGACATGAACAGACAATTCACAAAAAGANATANAAATGGCCCTTAAACATATGAAAAGATGTTCAACCTCACTCATAATAAGAGAAATGCAAATTAAAACTACACTGAGATACCATTTCTCACCTATCAGATTGGCAAAAATTCAAAAGTTTGACAACACACTCTGTTGGCGAGGCTGTGGGGAAACAGGCACTCTCATACATTGCTGGTGGGAATGCAAANTGGTACAACCCTTNTGGAGGGGAATTTGGCAATATCTAACAAAACTACATATGCATTTACCCTTTGACCCAGCAATCCCACTTCTAGGAATTTACCCTGAAGATACACCTCCAACATGTACAAAATNACATATGCACAAGGTTATTCATTGCAGCATTGTTTGTAATTGCAAAATATTGGAAACAACCTAAATGCCCATNNATAGGAGANTGGTTGAATAAACTATGGTACATCCACACAATGGAGTACTATGCAGCTGTAAAAAAGAATGAGGAAGATCTCTATGAACTGATATGGAGTGATTTCCAGGATATATTGTTAAGTGAAAAAAGCAAAGTGCAAAAGAGTATNTATAGTATGCTACCTTTCGTGTAAGAAAGAAGGGAAAATAAGAAAATATACATGTATCTGCTCATTTGTGCAAAAAGAAACACAGGAAGGATAAACCAGAAACTAATGAGATTGGTTACCTACAGGGGGTGGGTGGGAATGGGGAGAAAGAATGGGGGATGGGAACGGGATGGNAGGGATGAGGAGGGAGTGACACTTCTCTGAGTATACCTTTTTGTATAGTTCTGACTTTTAGAACCATGNTAATGTTTCACATACCTCAAAAATAAATAATTAAAATCAACCAGGATGTGGGGGGAACCCAAAATGGAATACAAACAGNAACAAATGAACCTAACTGTATTNCAAATGAATAACATAACCACACTGAAGGGGATGGGGAAGAAAAGAACTAACCTAAGTAACTTTGGAAAACAGTATTTTGACTGNATACTGTAAGGCTAAAGACAAAAAGAACTGTACACAAATACTGTACTCTAGTTAGTAAATTTGTTTCTCACAGGGGTATGGGTTAGCAATTCTGAAACTACTTTATGTGTATNCTAGGATTGAACAAATAAGTAAATATATTGTAGATAATGAGAGCCAGGTTTCTCACTGTCGGAGAAAGAAGTTACAAATAAGGAAAGGGGGAAGGCTAGAATGAACCCTGTGGTGTTGGATTGGAATCGGAGGTATCAGTATGAACTCATGGTTTTTAACATATATACAGATAGATAGATACAGAAATAAGATATAGATGTATGTGTATGCATGGGTTAGTATACATACATATATTTCCTAGCTCTGTCCGCTGAGAGGGCCTAGAAGCAATGACACCCCAGTAGCAATGAGCACACCTAGCGCCCAGATCTTGGTTTCTAAATACCATTCTCCAATAAAAGGAACCAGGGCTCCTTGGAGAAATGGCTGATTCCAGGGCTGGGGCAGGGAAAATACAAGATGAGCCTGGAACATCTTGTGGTGCCAGAAAGTAAGGAAGTGCTCAAAAAANGATGGGGGCATGTCAAAAGGACACAGGAGCCAACCTGAAGGAGCTCCCAATGGCCAAANCTGGAACAATTTGAGCAACAAAATAAATAATGATAGTANTGGATTATAACCCATAGAATAAAATAAATATCCATGAGTCCATACTGATATAAATAAATAATTGAATAAATAAATAAATGGGGGAGAAGGGACAGCTCTTCCTTACAGNAGAATTCCAATTAATAAATGTAGAAGGAATGANGGAAATAGAAAATCACCATTAGGCAAACACCACAGTAATAATTGTTGCAGGCAAGATCCACCGATGGATGCTAAAATTAGTGGGCGAAAGTTTGAGGAGAAACAGGATATTTGCATAGTCTCAAAGTATCTCCCCCAAGATATTTATTAATTACAAAGGGAAAAATAGTAACTTTACAGTGGAGAAACCCGGCAGACACCACCTTAACCAAGTGATCAAGGTTAACATCACCAGTAATAAGACATATCGACATCATGTACCCCCTGATATGATGCACTGAGAAGGGCACAACATCACTTCTGTGGTATTCTTGCCAAAAATGCATAACCTCAATCTAATCATGAGAAAACATCAGACAAACCCAAATTGAGGGACATTCTACAAAATAACTGACCAGTACTCTTCAAAAGTGTCAAGGTCATGAAAGACAAGGAAAGACTGAGGAACTGTCACAGATTGGAGGAGACTAAGGAGACATGACAACTAAATGCAATGTGGGATCCTGGATTGGATCCTGGAACAGAAAAAAGGACATTAGTGGAAAAACTGGTGAAATTCGAATAAGGTCTGTAGTTTAGTTAATAGTATTGTACCAATGTTAATTTCCTGGTTTTGATAATTGTACTATGGTTATGTAAGATGTTAACATTAGGGGAAGCTGGGTGAAGGGTATACGGGAACTCTCTGTACTATTTTTGCAACTTTTCTGTAAGTCTAAAATTATTTCAAAATAAAAAGTTAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MC4a_3end | ZNF281 | 803 | 812 | + | 16.91 | GGGGGATGGG |
| L1MC4a_3end | odd | 1469 | 1479 | + | 16.53 | CCCAGTAGCAA |
| L1MC4a_3end | AGL16 | 1263 | 1277 | - | 16.47 | TTTCCTTATTTGTAA |
| L1MC4a_3end | NR2F1 | 356 | 368 | - | 16.36 | GGGTCAAAGGGTA |
| L1MC4a_3end | ESRRA | 2296 | 2304 | + | 16.33 | TCAAGGTCA |
| L1MC4a_3end | Hnf4 | 356 | 368 | - | 16.26 | GGGTCAAAGGGTA |
| L1MC4a_3end | Nr5A2 | 2296 | 2304 | - | 16.13 | TGACCTTGA |
| L1MC4a_3end | SPL8B | 542 | 550 | - | 16.11 | ATAGTACTC |
| L1MC4a_3end | ZIPIC | 1565 | 1574 | - | 16.07 | CCAGCCCTGG |
| L1MC4a_3end | AP1 | 1265 | 1277 | + | 15.93 | ACAAATAAGGAAA |
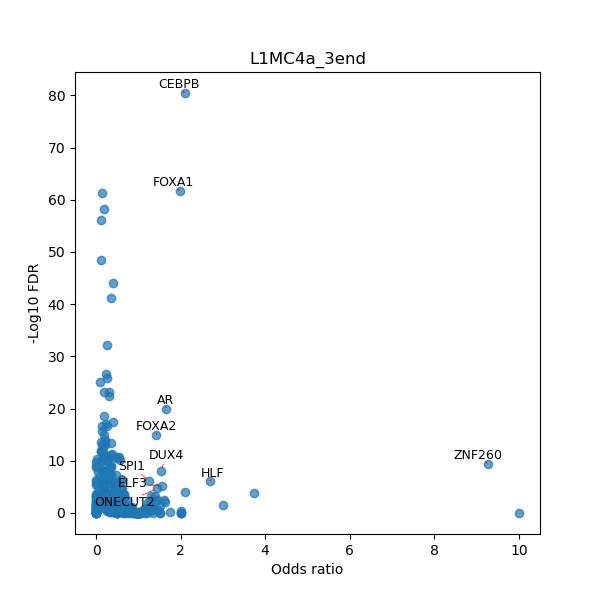
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.