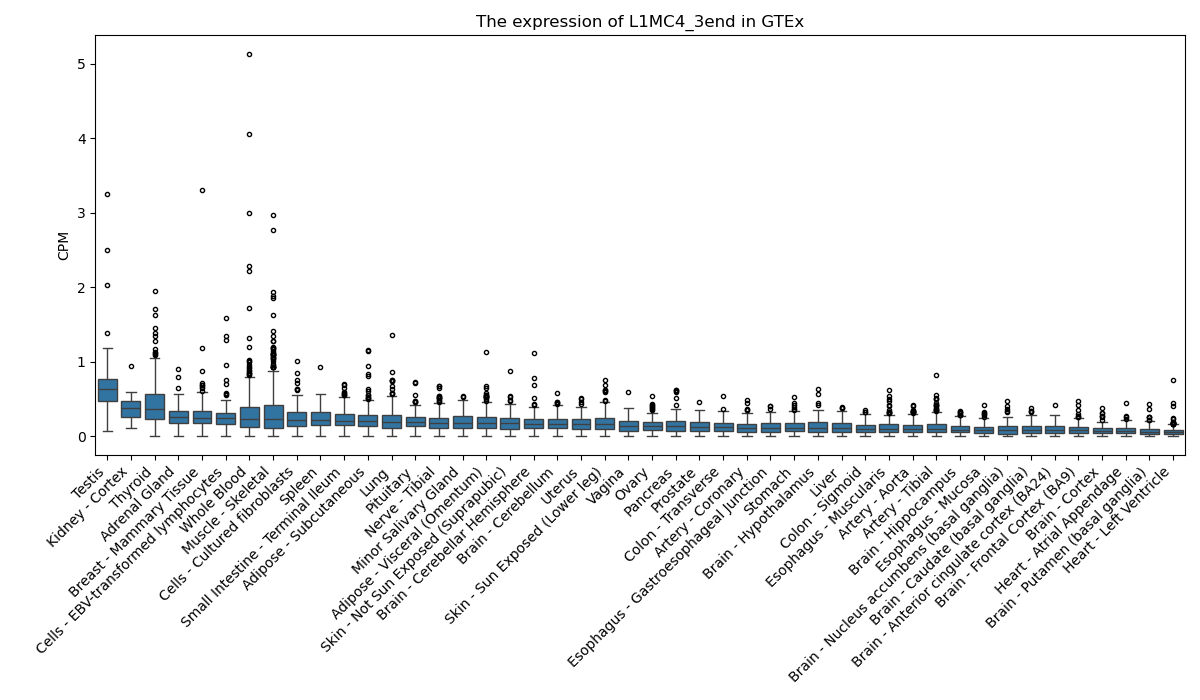
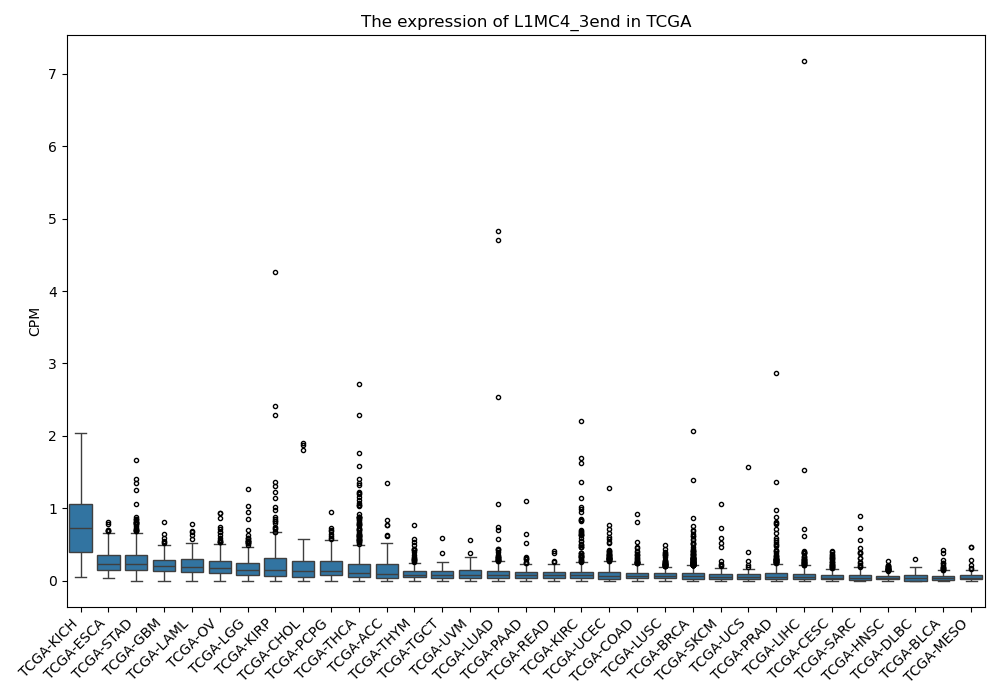
L1MC4_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000010 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2789 |
| Kimura value | 25.26 |
| Tau index | 0.7597 |
| Description | 3' end of L1 retrotransposon, L1MC4_3end subfamily |
| Comment | - |
| Sequence |
CTTGTATCCAGAATATATAAAGAACTCTTAAAACTCAACAATAAAAAAACAAACAACCCAATTAAAAAATGGGCAAAAGATTTGAACAGACATTTCACCAAAGAAGATATACAGATGGCAAATAAGCACATGAAAAGATGCTCAACATCATTNGTCATTAGGGAAATGCAAATTAAAACCACAATGAGATACCACTACACACCTATTAGAATGGCTAAAATCAAAAANACTGACAATACCAAATGCTGGCGAGGATGTGGAGCAACNGGAACTCTCATACATTGCTGGTGGGAATGCAAAATGGTACAGCCACTTTGGAAAACAGTTTGGCAGTTTCTTANAAAGCTAAACATACANTTACCATATGACCCAGCAATCNCACTCCTAGGTATTTACCCAAGNGAANTGAAAACNTATGTCCACACAAAAACCTGTACACGAATGTTTATAGCAGCTTTATTCATAATNGCCAAAANCTGGAAACAACCNAGATGTCCTTCAACAGGTGAATGGATAAACAAACTGTGGTACATCCATACAATGGAATACTACTCAGCGATAAAAAGGAATGAACTACTGATNCACGCAANAACATGGATGAATCTTAAATGCATNTTGCTAAGTGAAAGAAGCCAGNCNNAAAAGGCTACATACTGTATGATTCCATTTATATGACATTCTGGAAAAGGCAAAACTATAGAGACGGAAAACAGATCAGTGGTTGCCAGGGGTTGGGAGATGGGAAGNGGGGATGACTGCAAAGGNGAAGCACAGGGGATTTTTTAGGGTGATGAAACTATTCTGTATNAANTNNTCTGTATGATACTGTGGTGGTGGATACACGACNCTATGCATTTGTCAAAACCCATAGAACTTGTNANNANCCACAGANNTNTACAGCACAAAGAGTGAACTTTAATGTATGCAAATTTTAAAAAATCANTTAGGAGGTCGGGGGATCCCAGGATGGAATGCAGACTGTGACAAANGAATCTAACTGTATTACAAATGNATGANACAACCNCACTGAAGGGGATGGGGGAAAAAGGNGCTGACCTAAGTAACTTTGGAAATGAGTGGANNCTGTAAGGCTAAAGACAAAAAGAACTGTAAACAAACATTGAACTCTAGTTAGTAGGTTTGTTTTTCGCAGNGGTATGGGTTAGCAATTCTGAAACTACTTTCTGTACATNTTATACATTCTAGGATTGAGCAAATAAGTAAATATATTGNGGATAATGGGAGCCAGGTTTCTCACTGTCGGAGAAGGGAGTTACAAATATGGAAAGGGGGAAGGCTAGAATGAACCCTGTGGTGTTGGATTGGAATTGGAGGTATCGGTATGAACTCATGGTTTTTAATATATATGNATATACAGACGGACAGATATAGAAATAGATATAGATATATATGTGTNTGTGTATATGTGTATGTATATACGTACATATATTTCCTAGCTCTGTCCGCTGAGAGGGCCTAGAAGCAATGACACCCCAGTAGCAATGAGCACACCTAGCGCCCAGATCTTGGTTTCTAAATACCATTCTCCACTAAAAGGAACCAGGGCTCCTTGGAGAAATGGCTGATTCCAGGGCTGGGGCAGGGAAAGTACAAGATGAGCCTGGAACATCTTGTTGTGCCAGAAAGTAAGGAAGTGCTCAAAGAATGATGGGGACATGTCAAAAGGACACAGGAGCCAGCTTGAAGGGGCTCCCACTGGCCAAATCTGGGACAATTTGAGCATCAAAATAAATAATGATAGTAATGGATTATAACCCATTGAATAAAATAAGAATCCATGAGTCCATACTGATATAAATAAATAAATAAATAAATGGGGGAGAAGGGAAAGCTCTTCCTTACAGTAGAATGCCAACTAATAAATGTAGAAGGAATGATGGAATTAGAAAAATCACCATTTGGCAACCATCATAGTAATAATTGATTCAGGCAAGAATCATCAATGGATGCTAAAACTAGTGGGTGAAAGTTTGATGAGGAACAGGATATTTACATAGTCTCAAAGTATCTCCCCACAAANTACTTATTAATTACAAAGGGGAAAATAGTAACTTTACAGTGGAGAAACCTGGCAGACACCACCTTAACCAAGTGATCAAAGTTAACATCACCAGTAATGGGACAAATCGACATCATGTGCCTCCTGATATGATGCACTGAGAAGGACACAGCATCACTTCTGTGGTATTCCTGCCAAAAATGCATAACCTGAATCTAATCATGAGGAAACATCAGACAAACCCAAATTGAGGGACATTCTACAAAATAACTGGCCTGTACTCTTCAAAAATGTCAANGTCATGAAAGACAAAGAAAGACTGAGGAACTGTTCCAGATTAAAGGAGACTAAAGAGACATGACAACTAAATGCAACGCGTGATCCTGGATTGGATCCTGGACCAGANNNNNNNTTTGCTATAAAGGACATTATTGGGACAATTGGCGAAATTTGAATANGGTCTGTAGATTAGATAATAGTATTGTATCAATGTTAATTTCCTGATTTTGATAATTGTACTGTGGTTATGTAAGAGAATGTCCTTGTTCTTAGGAAATACACACTGAAGTATTTAGGGGTAAAGGGGCATNATGTCTGCAACTTACTCTCAAATGGTTCAGAAAAAAAAATATGTATATGCANANAGAGAATGATAAAGCAAATGTGGCAAAATGTTAACANTTGGNGAATCTGGGTGAAGGGTATACGGGAGTTCTTTGTACTATTCTTGCAACTTTTCTGTAAGTTTGAAATTATTTCAAAATAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MC4_3end | Bcl11B | 176 | 184 | + | 15.12 | AAACCACAA |
| L1MC4_3end | Foxj3 | 1110 | 1118 | + | 15.06 | GTAAACAAA |
| L1MC4_3end | nub | 922 | 932 | + | 14.99 | TATGCAAATTT |
| L1MC4_3end | AGL63 | 1274 | 1288 | + | 14.98 | TTACAAATATGGAAA |
| L1MC4_3end | POU2F1 | 922 | 930 | + | 14.94 | TATGCAAAT |
| L1MC4_3end | AP3 | 1276 | 1288 | + | 14.93 | ACAAATATGGAAA |
| L1MC4_3end | Nfe2l2 | 365 | 375 | + | 14.91 | ATGACCCAGCA |
| L1MC4_3end | TCX6 | 923 | 937 | + | 14.88 | ATGCAAATTTTAAAA |
| L1MC4_3end | ARF36 | 1670 | 1681 | + | 14.87 | TGGGGACATGTC |
| L1MC4_3end | POU3F2 | 921 | 932 | + | 14.87 | GTATGCAAATTT |
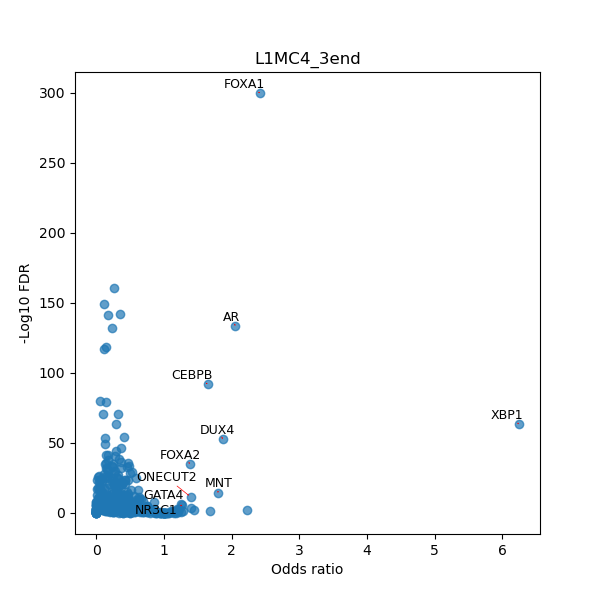
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.