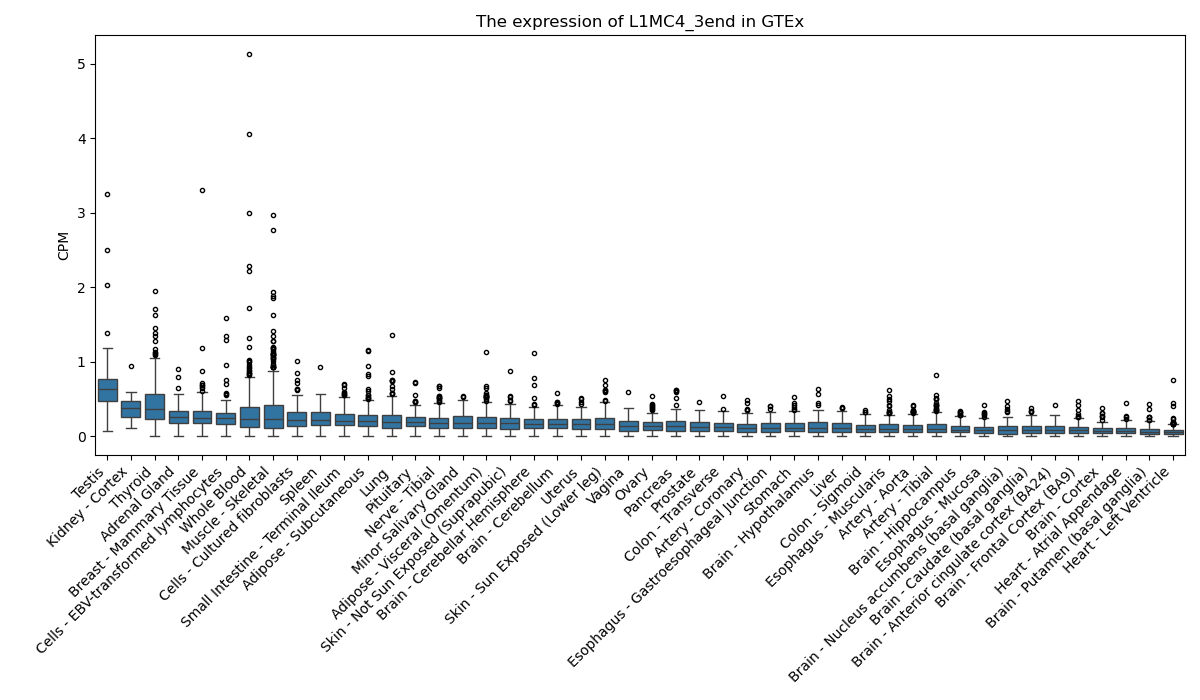
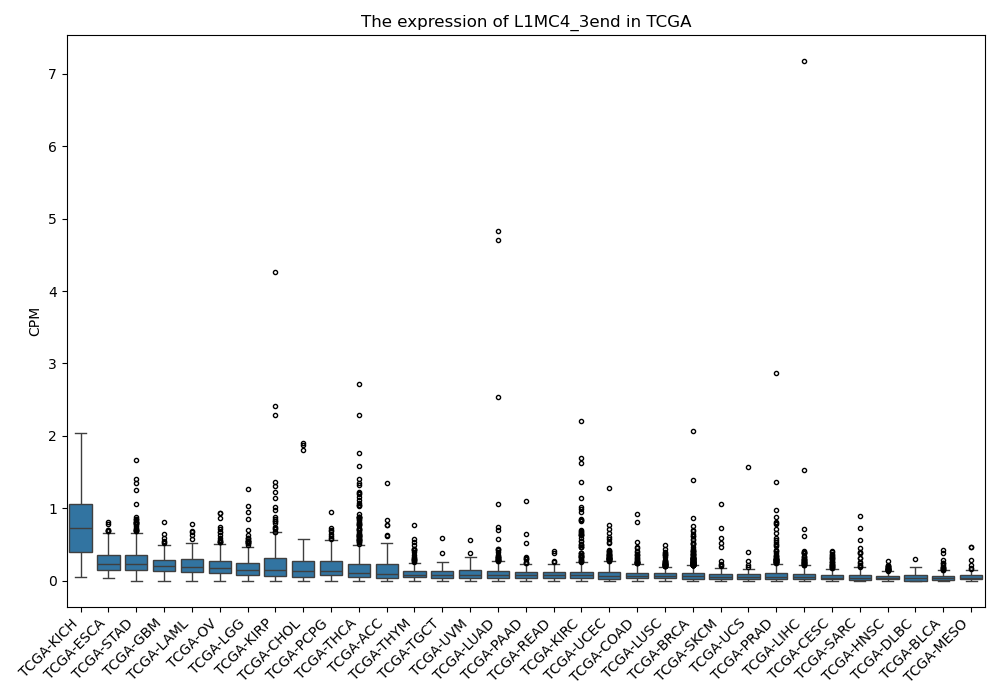
L1MC4_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000010 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2789 |
| Kimura value | 25.26 |
| Tau index | 0.7597 |
| Description | 3' end of L1 retrotransposon, L1MC4_3end subfamily |
| Comment | - |
| Sequence |
CTTGTATCCAGAATATATAAAGAACTCTTAAAACTCAACAATAAAAAAACAAACAACCCAATTAAAAAATGGGCAAAAGATTTGAACAGACATTTCACCAAAGAAGATATACAGATGGCAAATAAGCACATGAAAAGATGCTCAACATCATTNGTCATTAGGGAAATGCAAATTAAAACCACAATGAGATACCACTACACACCTATTAGAATGGCTAAAATCAAAAANACTGACAATACCAAATGCTGGCGAGGATGTGGAGCAACNGGAACTCTCATACATTGCTGGTGGGAATGCAAAATGGTACAGCCACTTTGGAAAACAGTTTGGCAGTTTCTTANAAAGCTAAACATACANTTACCATATGACCCAGCAATCNCACTCCTAGGTATTTACCCAAGNGAANTGAAAACNTATGTCCACACAAAAACCTGTACACGAATGTTTATAGCAGCTTTATTCATAATNGCCAAAANCTGGAAACAACCNAGATGTCCTTCAACAGGTGAATGGATAAACAAACTGTGGTACATCCATACAATGGAATACTACTCAGCGATAAAAAGGAATGAACTACTGATNCACGCAANAACATGGATGAATCTTAAATGCATNTTGCTAAGTGAAAGAAGCCAGNCNNAAAAGGCTACATACTGTATGATTCCATTTATATGACATTCTGGAAAAGGCAAAACTATAGAGACGGAAAACAGATCAGTGGTTGCCAGGGGTTGGGAGATGGGAAGNGGGGATGACTGCAAAGGNGAAGCACAGGGGATTTTTTAGGGTGATGAAACTATTCTGTATNAANTNNTCTGTATGATACTGTGGTGGTGGATACACGACNCTATGCATTTGTCAAAACCCATAGAACTTGTNANNANCCACAGANNTNTACAGCACAAAGAGTGAACTTTAATGTATGCAAATTTTAAAAAATCANTTAGGAGGTCGGGGGATCCCAGGATGGAATGCAGACTGTGACAAANGAATCTAACTGTATTACAAATGNATGANACAACCNCACTGAAGGGGATGGGGGAAAAAGGNGCTGACCTAAGTAACTTTGGAAATGAGTGGANNCTGTAAGGCTAAAGACAAAAAGAACTGTAAACAAACATTGAACTCTAGTTAGTAGGTTTGTTTTTCGCAGNGGTATGGGTTAGCAATTCTGAAACTACTTTCTGTACATNTTATACATTCTAGGATTGAGCAAATAAGTAAATATATTGNGGATAATGGGAGCCAGGTTTCTCACTGTCGGAGAAGGGAGTTACAAATATGGAAAGGGGGAAGGCTAGAATGAACCCTGTGGTGTTGGATTGGAATTGGAGGTATCGGTATGAACTCATGGTTTTTAATATATATGNATATACAGACGGACAGATATAGAAATAGATATAGATATATATGTGTNTGTGTATATGTGTATGTATATACGTACATATATTTCCTAGCTCTGTCCGCTGAGAGGGCCTAGAAGCAATGACACCCCAGTAGCAATGAGCACACCTAGCGCCCAGATCTTGGTTTCTAAATACCATTCTCCACTAAAAGGAACCAGGGCTCCTTGGAGAAATGGCTGATTCCAGGGCTGGGGCAGGGAAAGTACAAGATGAGCCTGGAACATCTTGTTGTGCCAGAAAGTAAGGAAGTGCTCAAAGAATGATGGGGACATGTCAAAAGGACACAGGAGCCAGCTTGAAGGGGCTCCCACTGGCCAAATCTGGGACAATTTGAGCATCAAAATAAATAATGATAGTAATGGATTATAACCCATTGAATAAAATAAGAATCCATGAGTCCATACTGATATAAATAAATAAATAAATAAATGGGGGAGAAGGGAAAGCTCTTCCTTACAGTAGAATGCCAACTAATAAATGTAGAAGGAATGATGGAATTAGAAAAATCACCATTTGGCAACCATCATAGTAATAATTGATTCAGGCAAGAATCATCAATGGATGCTAAAACTAGTGGGTGAAAGTTTGATGAGGAACAGGATATTTACATAGTCTCAAAGTATCTCCCCACAAANTACTTATTAATTACAAAGGGGAAAATAGTAACTTTACAGTGGAGAAACCTGGCAGACACCACCTTAACCAAGTGATCAAAGTTAACATCACCAGTAATGGGACAAATCGACATCATGTGCCTCCTGATATGATGCACTGAGAAGGACACAGCATCACTTCTGTGGTATTCCTGCCAAAAATGCATAACCTGAATCTAATCATGAGGAAACATCAGACAAACCCAAATTGAGGGACATTCTACAAAATAACTGGCCTGTACTCTTCAAAAATGTCAANGTCATGAAAGACAAAGAAAGACTGAGGAACTGTTCCAGATTAAAGGAGACTAAAGAGACATGACAACTAAATGCAACGCGTGATCCTGGATTGGATCCTGGACCAGANNNNNNNTTTGCTATAAAGGACATTATTGGGACAATTGGCGAAATTTGAATANGGTCTGTAGATTAGATAATAGTATTGTATCAATGTTAATTTCCTGATTTTGATAATTGTACTGTGGTTATGTAAGAGAATGTCCTTGTTCTTAGGAAATACACACTGAAGTATTTAGGGGTAAAGGGGCATNATGTCTGCAACTTACTCTCAAATGGTTCAGAAAAAAAAATATGTATATGCANANAGAGAATGATAAAGCAAATGTGGCAAAATGTTAACANTTGGNGAATCTGGGTGAAGGGTATACGGGAGTTCTTTGTACTATTCTTGCAACTTTTCTGTAAGTTTGAAATTATTTCAAAATAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MC4_3end | ZNF8 | 520 | 539 | - | 31.88 | GTATGGATGTACCACAGTTT |
| L1MC4_3end | ZNF135 | 944 | 957 | - | 18.46 | CCCCGACCTCCTAA |
| L1MC4_3end | PRDM9 | 1029 | 1048 | + | 18.21 | GAAGGGGATGGGGGAAAAAG |
| L1MC4_3end | SCRT1 | 499 | 508 | + | 17.58 | TCAACAGGTG |
| L1MC4_3end | FKH1 | 1106 | 1118 | + | 17.48 | AACTGTAAACAAA |
| L1MC4_3end | SOL1 | 923 | 937 | - | 16.97 | TTTTAAAATTTGCAT |
| L1MC4_3end | CG7928 | 1588 | 1599 | - | 16.92 | CCCAGCCCTGGA |
| L1MC4_3end | CHES-1-like | 1109 | 1118 | + | 16.77 | TGTAAACAAA |
| L1MC4_3end | CG12605 | 500 | 508 | - | 16.69 | CACCTGTTG |
| L1MC4_3end | squamosa | 1276 | 1288 | + | 16.53 | ACAAATATGGAAA |
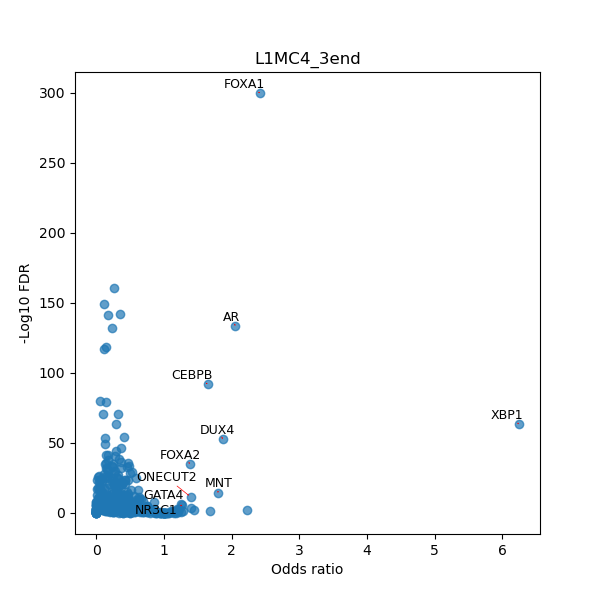
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.