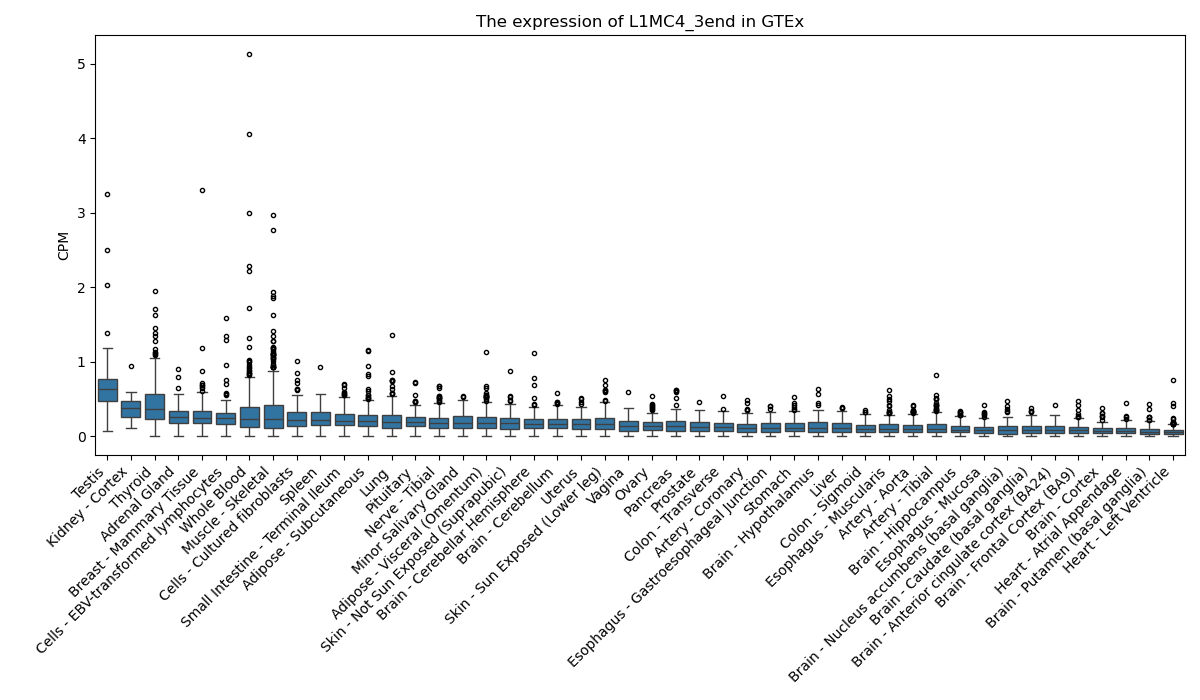
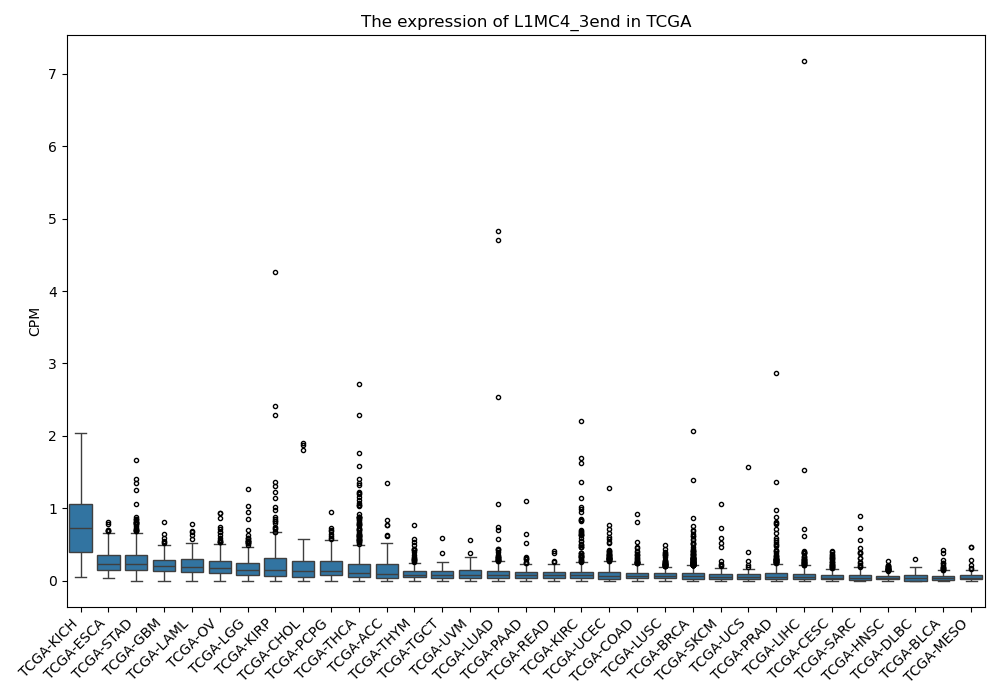
L1MC4_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000010 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2789 |
| Kimura value | 25.26 |
| Tau index | 0.7597 |
| Description | 3' end of L1 retrotransposon, L1MC4_3end subfamily |
| Comment | - |
| Sequence |
CTTGTATCCAGAATATATAAAGAACTCTTAAAACTCAACAATAAAAAAACAAACAACCCAATTAAAAAATGGGCAAAAGATTTGAACAGACATTTCACCAAAGAAGATATACAGATGGCAAATAAGCACATGAAAAGATGCTCAACATCATTNGTCATTAGGGAAATGCAAATTAAAACCACAATGAGATACCACTACACACCTATTAGAATGGCTAAAATCAAAAANACTGACAATACCAAATGCTGGCGAGGATGTGGAGCAACNGGAACTCTCATACATTGCTGGTGGGAATGCAAAATGGTACAGCCACTTTGGAAAACAGTTTGGCAGTTTCTTANAAAGCTAAACATACANTTACCATATGACCCAGCAATCNCACTCCTAGGTATTTACCCAAGNGAANTGAAAACNTATGTCCACACAAAAACCTGTACACGAATGTTTATAGCAGCTTTATTCATAATNGCCAAAANCTGGAAACAACCNAGATGTCCTTCAACAGGTGAATGGATAAACAAACTGTGGTACATCCATACAATGGAATACTACTCAGCGATAAAAAGGAATGAACTACTGATNCACGCAANAACATGGATGAATCTTAAATGCATNTTGCTAAGTGAAAGAAGCCAGNCNNAAAAGGCTACATACTGTATGATTCCATTTATATGACATTCTGGAAAAGGCAAAACTATAGAGACGGAAAACAGATCAGTGGTTGCCAGGGGTTGGGAGATGGGAAGNGGGGATGACTGCAAAGGNGAAGCACAGGGGATTTTTTAGGGTGATGAAACTATTCTGTATNAANTNNTCTGTATGATACTGTGGTGGTGGATACACGACNCTATGCATTTGTCAAAACCCATAGAACTTGTNANNANCCACAGANNTNTACAGCACAAAGAGTGAACTTTAATGTATGCAAATTTTAAAAAATCANTTAGGAGGTCGGGGGATCCCAGGATGGAATGCAGACTGTGACAAANGAATCTAACTGTATTACAAATGNATGANACAACCNCACTGAAGGGGATGGGGGAAAAAGGNGCTGACCTAAGTAACTTTGGAAATGAGTGGANNCTGTAAGGCTAAAGACAAAAAGAACTGTAAACAAACATTGAACTCTAGTTAGTAGGTTTGTTTTTCGCAGNGGTATGGGTTAGCAATTCTGAAACTACTTTCTGTACATNTTATACATTCTAGGATTGAGCAAATAAGTAAATATATTGNGGATAATGGGAGCCAGGTTTCTCACTGTCGGAGAAGGGAGTTACAAATATGGAAAGGGGGAAGGCTAGAATGAACCCTGTGGTGTTGGATTGGAATTGGAGGTATCGGTATGAACTCATGGTTTTTAATATATATGNATATACAGACGGACAGATATAGAAATAGATATAGATATATATGTGTNTGTGTATATGTGTATGTATATACGTACATATATTTCCTAGCTCTGTCCGCTGAGAGGGCCTAGAAGCAATGACACCCCAGTAGCAATGAGCACACCTAGCGCCCAGATCTTGGTTTCTAAATACCATTCTCCACTAAAAGGAACCAGGGCTCCTTGGAGAAATGGCTGATTCCAGGGCTGGGGCAGGGAAAGTACAAGATGAGCCTGGAACATCTTGTTGTGCCAGAAAGTAAGGAAGTGCTCAAAGAATGATGGGGACATGTCAAAAGGACACAGGAGCCAGCTTGAAGGGGCTCCCACTGGCCAAATCTGGGACAATTTGAGCATCAAAATAAATAATGATAGTAATGGATTATAACCCATTGAATAAAATAAGAATCCATGAGTCCATACTGATATAAATAAATAAATAAATAAATGGGGGAGAAGGGAAAGCTCTTCCTTACAGTAGAATGCCAACTAATAAATGTAGAAGGAATGATGGAATTAGAAAAATCACCATTTGGCAACCATCATAGTAATAATTGATTCAGGCAAGAATCATCAATGGATGCTAAAACTAGTGGGTGAAAGTTTGATGAGGAACAGGATATTTACATAGTCTCAAAGTATCTCCCCACAAANTACTTATTAATTACAAAGGGGAAAATAGTAACTTTACAGTGGAGAAACCTGGCAGACACCACCTTAACCAAGTGATCAAAGTTAACATCACCAGTAATGGGACAAATCGACATCATGTGCCTCCTGATATGATGCACTGAGAAGGACACAGCATCACTTCTGTGGTATTCCTGCCAAAAATGCATAACCTGAATCTAATCATGAGGAAACATCAGACAAACCCAAATTGAGGGACATTCTACAAAATAACTGGCCTGTACTCTTCAAAAATGTCAANGTCATGAAAGACAAAGAAAGACTGAGGAACTGTTCCAGATTAAAGGAGACTAAAGAGACATGACAACTAAATGCAACGCGTGATCCTGGATTGGATCCTGGACCAGANNNNNNNTTTGCTATAAAGGACATTATTGGGACAATTGGCGAAATTTGAATANGGTCTGTAGATTAGATAATAGTATTGTATCAATGTTAATTTCCTGATTTTGATAATTGTACTGTGGTTATGTAAGAGAATGTCCTTGTTCTTAGGAAATACACACTGAAGTATTTAGGGGTAAAGGGGCATNATGTCTGCAACTTACTCTCAAATGGTTCAGAAAAAAAAATATGTATATGCANANAGAGAATGATAAAGCAAATGTGGCAAAATGTTAACANTTGGNGAATCTGGGTGAAGGGTATACGGGAGTTCTTTGTACTATTCTTGCAACTTTTCTGTAAGTTTGAAATTATTTCAAAATAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MC4_3end | POU2F3 | 922 | 930 | + | 15.87 | TATGCAAAT |
| L1MC4_3end | AGL16 | 1274 | 1288 | - | 15.86 | TTTCCATATTTGTAA |
| L1MC4_3end | INSM1 | 723 | 734 | + | 15.83 | TGCCAGGGGTTG |
| L1MC4_3end | POU3F4 | 922 | 930 | + | 15.57 | TATGCAAAT |
| L1MC4_3end | ZNF816 | 1670 | 1684 | + | 15.47 | TGGGGACATGTCAAA |
| L1MC4_3end | POU3F3 | 920 | 932 | + | 15.42 | TGTATGCAAATTT |
| L1MC4_3end | SGR5 | 1091 | 1103 | + | 15.39 | GCTAAAGACAAAA |
| L1MC4_3end | FOXC1 | 2007 | 2017 | - | 15.30 | TATGTAAATAT |
| L1MC4_3end | FOXN3 | 1110 | 1117 | + | 15.19 | GTAAACAA |
| L1MC4_3end | FOXB1 | 1218 | 1228 | + | 15.15 | TAAGTAAATAT |
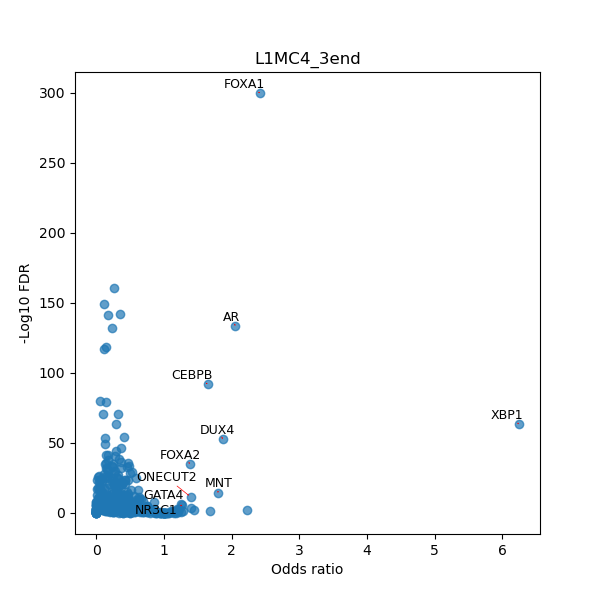
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.