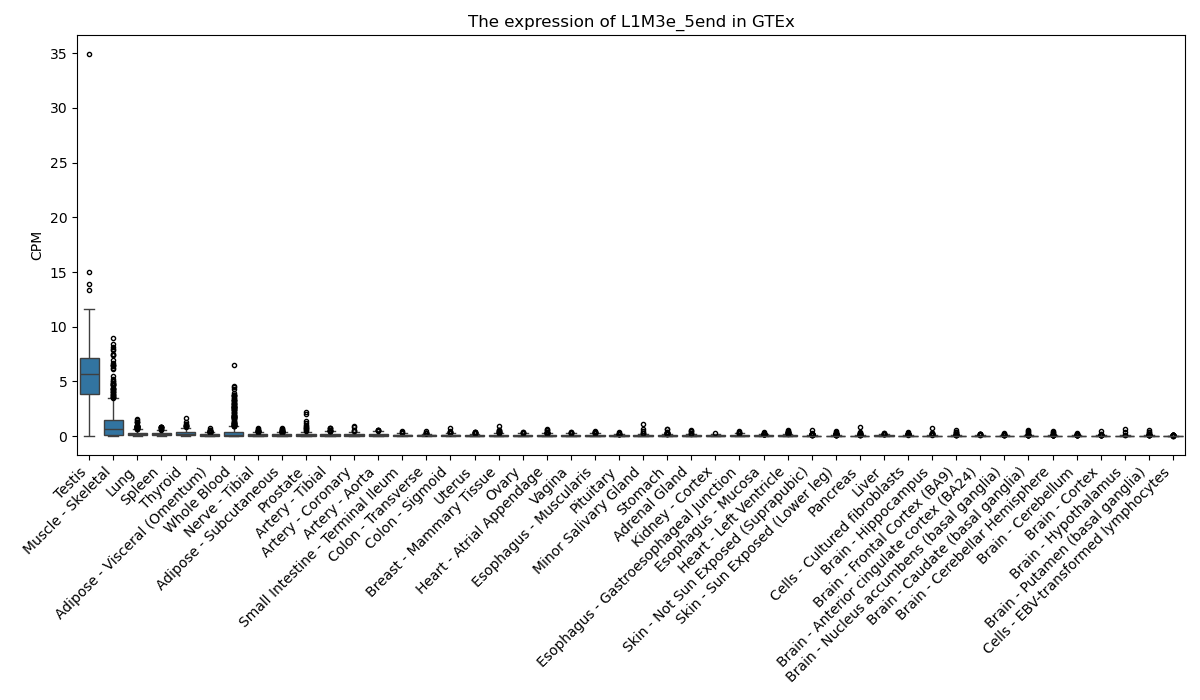
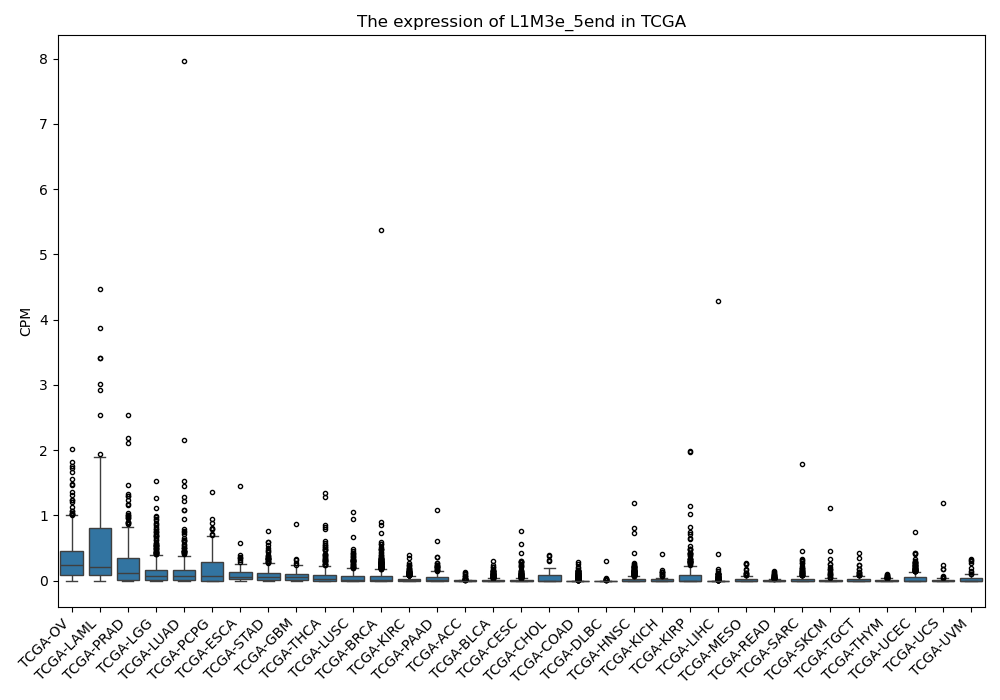
L1M3e_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000242 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Euarchontoglires |
| Length | 2113 |
| Kimura value | 24.19 |
| Tau index | 0.9892 |
| Description | 5' end of L1 retrotransposon, L1M3e_5end subfamily |
| Comment | Contains multiple ~30 bp tandem duplications in the promoter region. |
| Sequence |
GACGTCAGCAAGATGGCGGAATAGGACTTTCCAGCGCTCGTCCTCACAGAAACATCAATTTGAACAACTATCCACGCACGAAAATACCTTCACAAGAGCTAAGGAAACCAGGTGAGAGATTACAGCACCTGGGTGTAGCACAGAAATAAGAAAAGACGCATTGAAGAGGGTAGGAAGGACAGTTTTACATTACCCGCGTCACCCCTCCCCCAACCCCAGGCAGCACAGCGTGGAGAGAGATACCCTCCGCNTGGGGGAAGGAGAGGGAAGTGAGCACCGGACTTTGCCTCGGACCCCAANCACCGGGCCCGCCCCAGTAAAACCCAGCGCCGGGCAGGCCCCCACGGCCCCAGACTCCAGGCCGGTACCCGCGGACTGAGCCGCGCCAGATCCCACAGCCCAGGCTCCAGGCCTGCCCGGCGGACTCAGTCTCCAGGCCCGCCCCAGCGCCAGGCCGACCCCAGTGCCCCAGGCTCCAGACCGGCCCCAGCGCCAGGCCGGCCCCAGCGGCCCCAGGCTCCAGGCCGNCCCCAGCGCCAGGCCGGCCCCCGCGGCCCCAGGCTTCAGGCCCGCCCCAGCACCAGGCCGGCCCCCGCAGCCCTAGNCATCAGGCCAGCACCCGCGGACCCAGCCTCCAGGCCGGCCCCTGCGGACACAGGCTCCAGGCCTACCCAGCGCCAGGCCAGCCCCTGCGGCCCCAGGCTCCAGGCCCACCCCAGGTTCCAGACCAGCCCAGAGCCAGGTCGGCCCACGTAGCCCCAGGCTTCAGGCCCGCCCCAGCGCCAGGTCAGCACCCCTGGCCTCAGACCNCGNCCCGGTACCAGGCCGGCACCCGCGGACACAGGCTCCAGGCCTGCCCAGTGCCAGGCCGGTCCCTGCGGCCCCACCCTCCAGGNCCGGCCCCTGCGGCCCCACGCTCCAGCAGACCCAGGGTCCAGGCCCGTCCCAGTAGACCCCAGCGCTAGGCCGGCCCCCACGGACCCAGGCTCCAGGACCGCCCCTGTGGACCCAGGTCCCAGGCCGGCCCCCGCGGCCCCAGGACCCAGGCCAGCCCTCGNAGACCTAGCCTCCAGGCCAGCCCTGCAGACCCAGCCTCCAGGCCGGCNCCCGCGGACCCAGGCTCCAGGCCATCCCCCAGGCTCCAGGCCAGCCCCAGTGGCTCCAGGCACCAGGCCAGCNCCCGCGGACCCAGGCTCCAGACCGGCCCCACGCTACCCCAGCACCAGGCCAGCCCCAGGCTCCAGGCCGGTCCCCGTGGCCCAGGCTCCAGTGGACCCAGGGTCCAGGCCTGCTCCAGCAGACCCAGGGTCCAGGCCCACCCCAGTAGACCCCGGCGCCAGGCCGGCCCCCGCGGACCCAGGCTCCAGGACCACCCCTGCAGACCCAGGCTCCAGGCCAGCCCCCGCGGACCAGGATCCANGGCCCANNNCCNCAGNCGCAGGCTCCAGGCCCGCCCCAGCGCCAGGCCAGCCCCCATGGACTCAGGCTCCAGGCCCGTCCCAGTGGACCCAGGCNCCAGGCCCATCCCAGCGCCAGGCCGGCCCCTGCAGACTCAGGCTCAAGGCCCACCCCAGCGCCAGGTCAGCCCCTGTGGACCCAGGCTTCAGGCCGGCCCCCGCGGACNCAGGCTCCAGGCCCGCCCTCGTGGACCCAGGCTCCAGGCCCACCCCCGCGGACCCAATCAACAGGTCCACCCAGTGGATCCAGGCTCCAGGCCCAACCCCGCGGACCCAGGCGCCAGGCCCGCCACCTGCTGACCCAGGCACCAGGCCAGCCTGCCCGAGGACTCCAGCAGCAAGCCCGCCCGCGGACCACGCCAGACGGCCTGCCCAGAATCTCTGGATGACTGGTGAAGGGCTTTCCCNGACAAAGCCAGTCTGCAAAGACTGGAATAAGTNCCTACTTCTTCAAATGCGCAGACACCAACGCAAGGCCCAAGGATCACGAANAATCAGGGAAACATGACACCACCAAAGGAACAAAATAAAGCGCCAGTAACCGACCCTAAAGAAATGGAGATNTACGAACTGCCTGACAAAGAATTCAAAATAATCGTTTTAAGGAAGCTCAGTGAGCTNCAAGAGAACACAGATAAACAACTAAACGAAATCAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1M3e_5end | NFIB | 1162 | 1178 | - | 15.69 | GCTGGCCTGGTGCCTGG |
| L1M3e_5end | NFIB | 1759 | 1775 | - | 15.69 | GCTGGCCTGGTGCCTGG |
| L1M3e_5end | SP2 | 1449 | 1457 | - | 15.67 | GGGGCGGGC |
| L1M3e_5end | SP2 | 307 | 315 | - | 15.67 | GGGGCGGGC |
| L1M3e_5end | SP2 | 437 | 445 | - | 15.67 | GGGGCGGGC |
| L1M3e_5end | SP2 | 568 | 576 | - | 15.67 | GGGGCGGGC |
| L1M3e_5end | SP2 | 770 | 778 | - | 15.67 | GGGGCGGGC |
| L1M3e_5end | ZNF213 | 1258 | 1269 | + | 15.61 | GCCCAGGCTCCA |
| L1M3e_5end | ZNF213 | 398 | 409 | + | 15.61 | GCCCAGGCTCCA |
| L1M3e_5end | ZNF524 | 288 | 296 | + | 15.60 | CTCGGACCC |
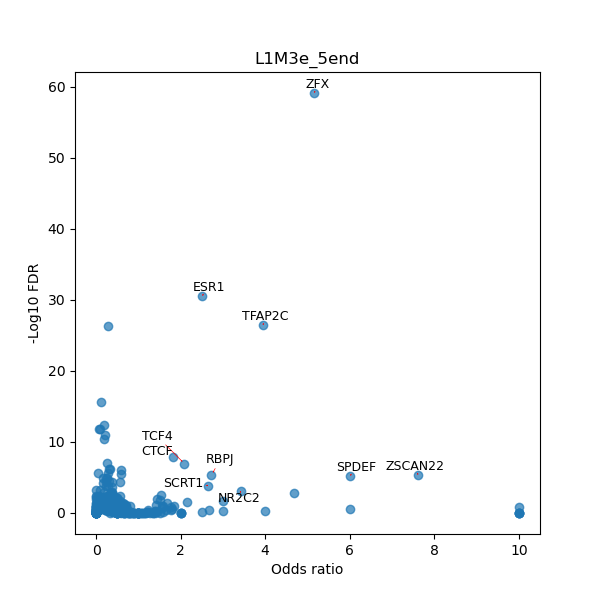
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.