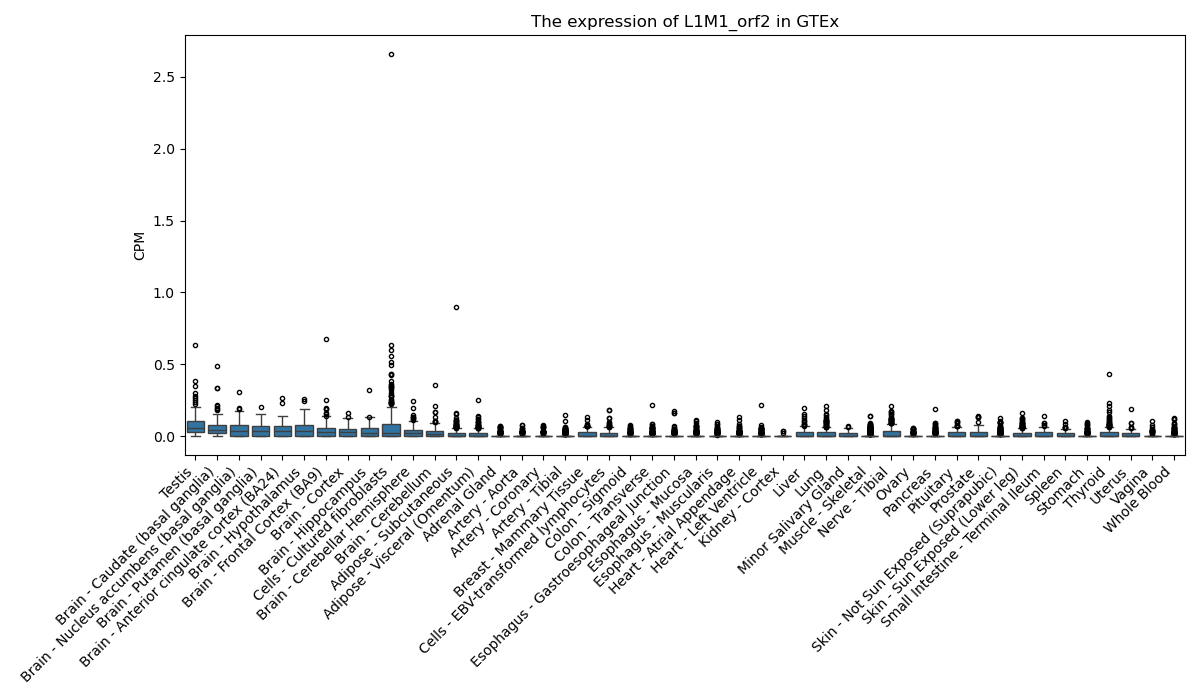
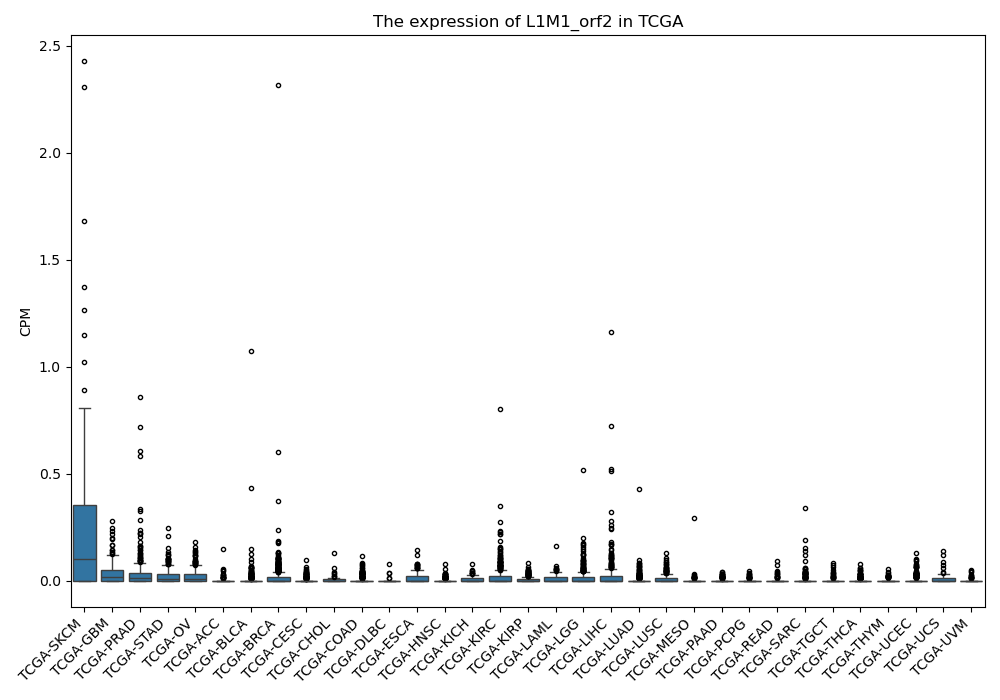
L1M1_orf2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000228 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Haplorrhini |
| Length | 3294 |
| Kimura value | 11.94 |
| Tau index | 0.8801 |
| Description | ORF2 from L1 retrotransposon, L1M1_orf2 subfamily |
| Comment | ORF2 from L1M1-like LINEs. |
| Sequence |
ATGGCAGGAGTAAGTCCTTACTTATCAATAATAACATTGAATGTAAATGGACTAAACTCTCCAATCAAAAGACATAGAGTGGCTGAATGGATGAAAAAACAAGACCCANTGATCTGTTGCCTACAAGAAACACACTTCACCTATAAAGACACACATAGACTGAAAATAAAGGGATGGAAAAAGATATTCCATGCCAATGGAAACCAAAAAAGAGCAGGAGTAGCTATACTTATATCAGACAAAATAGATTTCAAGACAAAAACTATAAGAAGAGACAAAGAAGGTCACTATATAATGATAAAGGGGTCAATTCAGCAAGAGGATATAACAATTNTAAATATATATGCACCCAACACTGGAGCACCCAGATATATAAAGCAAATATTATTAGAGCTAAAGAGAGAGATAGACCCCAATACAATAATAGCTGGAGACTTCAACACCCCACTTTCAGCATTGGACAGATCTTCCAGACAGAAAATCAACAAAGAAACATCGGACTTAATCTGCACTATAGACCAAATGGACCTAATAGATATTTACAGAACATTTCATCCAACGGCTGCAGAATACACATTCTTTTCCTCAGCACATGGATCATTCTCAAGGATAGACCATATGTTAGGTCACAAAACAAGTCTTAAAACATTCAAAAAAATTGAAATAATATCAAGCATCTTCTCTGACCACAATGGAATAAAACTAGAAATCAATAACAAGAGGAATTTTGGAAACTATACAAACACATGGAAATTAAACAATATGCTCCTGAATGACCAGTGGGTCAATGAAGAAATTAAGAAGGAAATTGAAAAATTTCTTGAAACAAATGATAATGGAAACACAACATACCAAAACCTATGGGATACAGCAAAAGCAGTACTAAGAGGGAAGTTTATAGCTATAAGTGCCTACATCAAAAAAGAAGAAAAACTTCAAATAAACAACCTAACGATGCATCTTAAAGAACTAGAAAAGCAAGAGCAAACCAAACCCAAAATTAGTAGAAGAAAAGAAATAATAAAGATCAGAGCAGAAATAAATGAAATTGAAATGAAGAAAACAATACAAAAGATCAATGAAACAAAAAGTTGGTTTTTTGAAAAGNTAAACAAAATTGACAAACCTTTAGCCAGACTAANNAAGAAAAAAAGAGAGAAGACCCAAATAAATAAAATCAGAGATGAAAAAGGAGACATTACAACTGATACCGCAGAAATTCAAAGGATCATTAGTGGCTACTATGAGCAACTATATGCCAATAAATTGGAAAATCTAGAAGAAATGGACAAATTCCTAGACACATACAACCTACCAAGATTGAACCATGAAGAAATCCAAAACCTGAACAGACCAATAACAAGTAACGAGATCGAAGCCGTAATAAAAAGTCTCCCAGTAAAGAAAAGCCCGGGACCCGATGGCTTCACTGCTGAATTCTACCAAACATTTAAAGAAGAACTAATACCAATCCTACTCAAACTATTCCGAAAAATAGAGGAGGAGGGAATACTTCCAAACTCATTCTACGAGGCCAGTATTACCCTGATACCAAAACCAGACAAAGACACATCAAAAAAAGAAAACTACAGGCCAATATCTCTGATGAATATTGATGCAAAAATCCTCAACAAAATACTAGCAAACCGAATTCAACAATACATTAAAAAGATCATTCATCATGACCAAGTGGGATTTATCCCNGGGATGCAAGGATGGTTCAACATACGCAAATCAATCAATGTGATACATCATATCAACAGAATGAAGGACAAAAACCATATGATCATTTCAATTGATGCTGAAAAAGCATTTGATAAAATTCAACATCCCTTCATGATAAAAACCCTCAAAAAACTGGGTATAGAAGGAACATACCTCAACATAATAAAAGCCATATACGACAGACCCACAGCTAGTATCATACTGAATGGGGAAAAACTGAAAGCCTTTCCTCTAAGATCTGGAACACGACAAGGATGCCCACTTTCACCACTGTTATTCAACATAGTACTGGAAGTCCTAGCTAGAGCAATCAGACAAGAGAAAGAAATAAAGGGCATCCAAATTGGAAAGGAAGAAGTCAAATTATCCTTGTTTGCAGATGATATGATCTTATATTTGGAAAAACCTAAAGACTCCACNAAAAAACTATTAGAACTGATAAACAAATTCAGTAAAGTTGCAGGATACAAAATCAACATACAAAAATCAGTAGCATTTCTATATGCCAACAGTGAACAATCTGAAAAAGAAATCAAAAAAGTAATCCCATTTACAATAGCCACAAATAAAATTAAATACCTAGGAATTAACTTAACCAAAGAAGTGAAAGATCTCTACAATGAAAACTATAAAACACTGATGAAAGAAATTGAAGAGGACACCAAAAAATGGAAAGATATTCCATGTTCATGGATTGGAAGAATCAATATTGTTAAAATGTCCATACTACCCAAAGCAATCTACAGATTCAATGCAATCCCTATCAAAATACCAATGACATTCTTCACAGAAATAGAAAAAACAATCCTAAAATTTATATGGAACCACAAAAGACCCAGAATAGCCAAAGCTATCCTGAGCAAAAAGAACAAAACTGGAGGAATCACATTACCTGACTTCAAATTATACTACAGAGCTATAGTAACCAAAACAGCATGGTACTGGCATAAAAACAGACACATAGACCAATGGAACAGAATAGAGAACCCAGAAACAAATCCACACACCTACAGTGAACTCATTTTCGACAAAGGTGCCAAGAACATACACTGGGGAAAAGACAGTCTCTTCAATAAATGGTGCTGGGAAAACTGGATATCCATATGCAGAAGAATGAAACTAGACCCCTATCTCTCGCCATATACAAAAATCAAATCAAAATGGATTAAAGACTTAAATCTAAGACCTCAAACTATGAAACTACTACAAGAAAACATTGGGGAAACTCTCCAGGACATTGGTCTGGGCAAAGATTTCTTGAGTAATACCCCACAAGCACAGGCAACCAAAGCAAAAATGGACAAATGGGATCACATCAAGTTAAAAAGCTTCTGCACAGCAAAGGAAACAATCAACAAAGTGAAGAGACAACCCACAGAATGGGAGAAAATATTTGCAAACTACCCATCTGACAAGGGATTAATAACCAGAATATATAAGGAGCTCAAACAACTCTATAGGAAAAAATCTAATAATCCGATTAAAAAATGGGCAAAAGATCTGAATAGACATTTCTCAAAAGAAGACATACAAATGGCAAACAGGCATATGAAAAGGTGCTCAACATCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1M1_orf2 | TCX6 | 641 | 655 | - | 16.27 | TTTTGAATGTTTTAA |
| L1M1_orf2 | DOF3.6 | 2250 | 2270 | - | 16.14 | ATTACTTTTTTGATTTCTTTT |
| L1M1_orf2 | AGL27 | 2063 | 2076 | - | 16.13 | CTTTCCAATTTGGA |
| L1M1_orf2 | OLIG1 | 614 | 623 | + | 16.04 | ACCATATGTT |
| L1M1_orf2 | PBX1 | 1734 | 1742 | + | 15.97 | ATCAATCAA |
| L1M1_orf2 | dimm | 614 | 623 | - | 15.87 | AACATATGGT |
| L1M1_orf2 | dimm | 614 | 623 | + | 15.79 | ACCATATGTT |
| L1M1_orf2 | DOF3.6 | 2249 | 2269 | - | 15.68 | TTACTTTTTTGATTTCTTTTT |
| L1M1_orf2 | OLIG2 | 614 | 623 | + | 15.67 | ACCATATGTT |
| L1M1_orf2 | Ddit3::Cebpa | 2477 | 2486 | + | 15.67 | ATGCAATCCC |
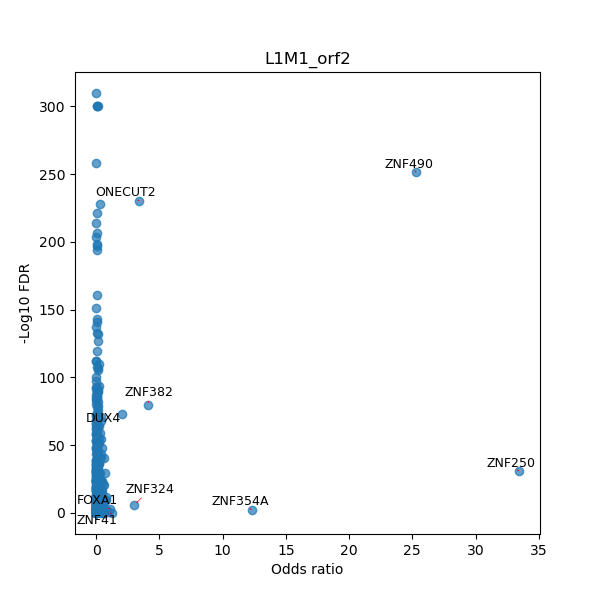
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.