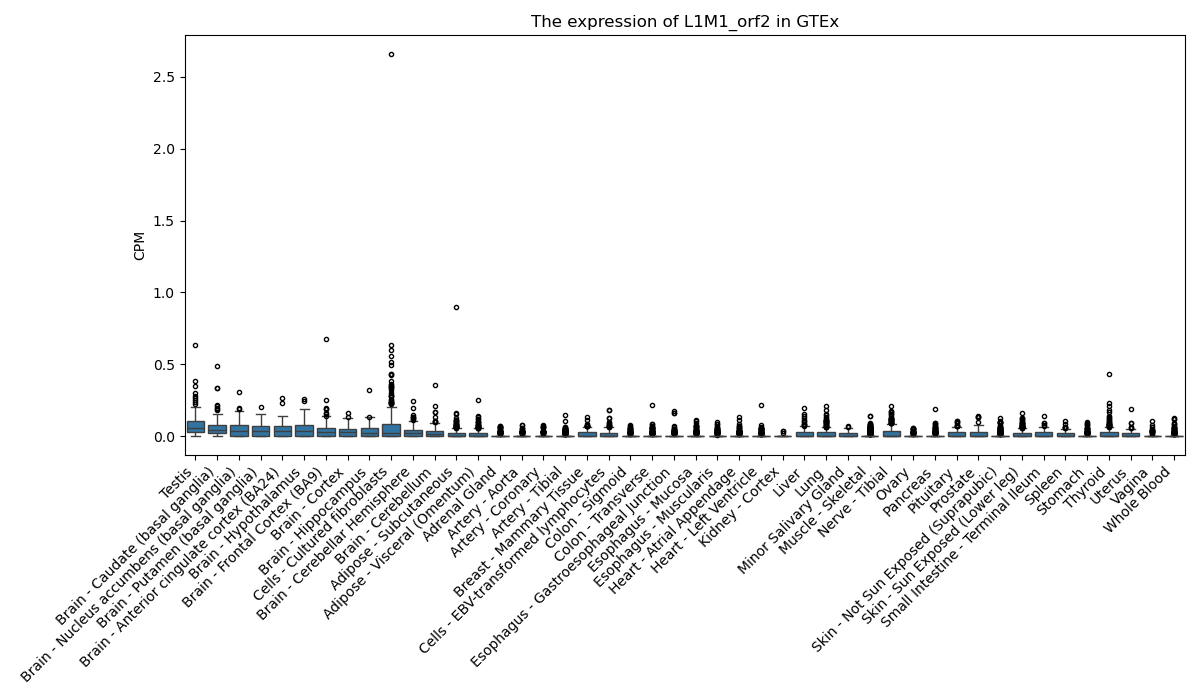
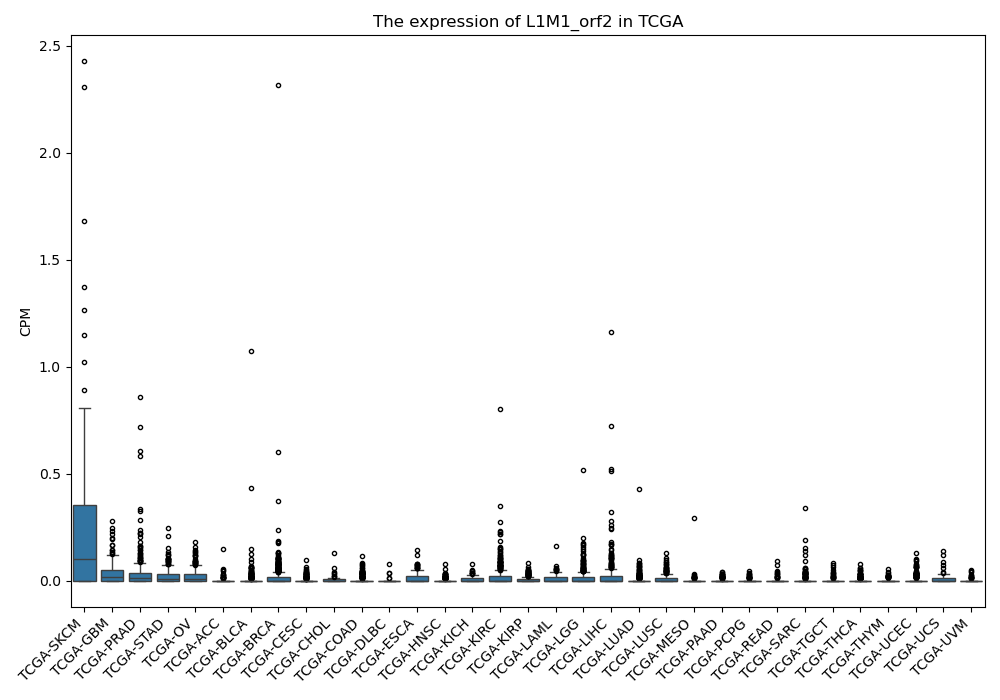
L1M1_orf2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000228 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Haplorrhini |
| Length | 3294 |
| Kimura value | 11.94 |
| Tau index | 0.8801 |
| Description | ORF2 from L1 retrotransposon, L1M1_orf2 subfamily |
| Comment | ORF2 from L1M1-like LINEs. |
| Sequence |
ATGGCAGGAGTAAGTCCTTACTTATCAATAATAACATTGAATGTAAATGGACTAAACTCTCCAATCAAAAGACATAGAGTGGCTGAATGGATGAAAAAACAAGACCCANTGATCTGTTGCCTACAAGAAACACACTTCACCTATAAAGACACACATAGACTGAAAATAAAGGGATGGAAAAAGATATTCCATGCCAATGGAAACCAAAAAAGAGCAGGAGTAGCTATACTTATATCAGACAAAATAGATTTCAAGACAAAAACTATAAGAAGAGACAAAGAAGGTCACTATATAATGATAAAGGGGTCAATTCAGCAAGAGGATATAACAATTNTAAATATATATGCACCCAACACTGGAGCACCCAGATATATAAAGCAAATATTATTAGAGCTAAAGAGAGAGATAGACCCCAATACAATAATAGCTGGAGACTTCAACACCCCACTTTCAGCATTGGACAGATCTTCCAGACAGAAAATCAACAAAGAAACATCGGACTTAATCTGCACTATAGACCAAATGGACCTAATAGATATTTACAGAACATTTCATCCAACGGCTGCAGAATACACATTCTTTTCCTCAGCACATGGATCATTCTCAAGGATAGACCATATGTTAGGTCACAAAACAAGTCTTAAAACATTCAAAAAAATTGAAATAATATCAAGCATCTTCTCTGACCACAATGGAATAAAACTAGAAATCAATAACAAGAGGAATTTTGGAAACTATACAAACACATGGAAATTAAACAATATGCTCCTGAATGACCAGTGGGTCAATGAAGAAATTAAGAAGGAAATTGAAAAATTTCTTGAAACAAATGATAATGGAAACACAACATACCAAAACCTATGGGATACAGCAAAAGCAGTACTAAGAGGGAAGTTTATAGCTATAAGTGCCTACATCAAAAAAGAAGAAAAACTTCAAATAAACAACCTAACGATGCATCTTAAAGAACTAGAAAAGCAAGAGCAAACCAAACCCAAAATTAGTAGAAGAAAAGAAATAATAAAGATCAGAGCAGAAATAAATGAAATTGAAATGAAGAAAACAATACAAAAGATCAATGAAACAAAAAGTTGGTTTTTTGAAAAGNTAAACAAAATTGACAAACCTTTAGCCAGACTAANNAAGAAAAAAAGAGAGAAGACCCAAATAAATAAAATCAGAGATGAAAAAGGAGACATTACAACTGATACCGCAGAAATTCAAAGGATCATTAGTGGCTACTATGAGCAACTATATGCCAATAAATTGGAAAATCTAGAAGAAATGGACAAATTCCTAGACACATACAACCTACCAAGATTGAACCATGAAGAAATCCAAAACCTGAACAGACCAATAACAAGTAACGAGATCGAAGCCGTAATAAAAAGTCTCCCAGTAAAGAAAAGCCCGGGACCCGATGGCTTCACTGCTGAATTCTACCAAACATTTAAAGAAGAACTAATACCAATCCTACTCAAACTATTCCGAAAAATAGAGGAGGAGGGAATACTTCCAAACTCATTCTACGAGGCCAGTATTACCCTGATACCAAAACCAGACAAAGACACATCAAAAAAAGAAAACTACAGGCCAATATCTCTGATGAATATTGATGCAAAAATCCTCAACAAAATACTAGCAAACCGAATTCAACAATACATTAAAAAGATCATTCATCATGACCAAGTGGGATTTATCCCNGGGATGCAAGGATGGTTCAACATACGCAAATCAATCAATGTGATACATCATATCAACAGAATGAAGGACAAAAACCATATGATCATTTCAATTGATGCTGAAAAAGCATTTGATAAAATTCAACATCCCTTCATGATAAAAACCCTCAAAAAACTGGGTATAGAAGGAACATACCTCAACATAATAAAAGCCATATACGACAGACCCACAGCTAGTATCATACTGAATGGGGAAAAACTGAAAGCCTTTCCTCTAAGATCTGGAACACGACAAGGATGCCCACTTTCACCACTGTTATTCAACATAGTACTGGAAGTCCTAGCTAGAGCAATCAGACAAGAGAAAGAAATAAAGGGCATCCAAATTGGAAAGGAAGAAGTCAAATTATCCTTGTTTGCAGATGATATGATCTTATATTTGGAAAAACCTAAAGACTCCACNAAAAAACTATTAGAACTGATAAACAAATTCAGTAAAGTTGCAGGATACAAAATCAACATACAAAAATCAGTAGCATTTCTATATGCCAACAGTGAACAATCTGAAAAAGAAATCAAAAAAGTAATCCCATTTACAATAGCCACAAATAAAATTAAATACCTAGGAATTAACTTAACCAAAGAAGTGAAAGATCTCTACAATGAAAACTATAAAACACTGATGAAAGAAATTGAAGAGGACACCAAAAAATGGAAAGATATTCCATGTTCATGGATTGGAAGAATCAATATTGTTAAAATGTCCATACTACCCAAAGCAATCTACAGATTCAATGCAATCCCTATCAAAATACCAATGACATTCTTCACAGAAATAGAAAAAACAATCCTAAAATTTATATGGAACCACAAAAGACCCAGAATAGCCAAAGCTATCCTGAGCAAAAAGAACAAAACTGGAGGAATCACATTACCTGACTTCAAATTATACTACAGAGCTATAGTAACCAAAACAGCATGGTACTGGCATAAAAACAGACACATAGACCAATGGAACAGAATAGAGAACCCAGAAACAAATCCACACACCTACAGTGAACTCATTTTCGACAAAGGTGCCAAGAACATACACTGGGGAAAAGACAGTCTCTTCAATAAATGGTGCTGGGAAAACTGGATATCCATATGCAGAAGAATGAAACTAGACCCCTATCTCTCGCCATATACAAAAATCAAATCAAAATGGATTAAAGACTTAAATCTAAGACCTCAAACTATGAAACTACTACAAGAAAACATTGGGGAAACTCTCCAGGACATTGGTCTGGGCAAAGATTTCTTGAGTAATACCCCACAAGCACAGGCAACCAAAGCAAAAATGGACAAATGGGATCACATCAAGTTAAAAAGCTTCTGCACAGCAAAGGAAACAATCAACAAAGTGAAGAGACAACCCACAGAATGGGAGAAAATATTTGCAAACTACCCATCTGACAAGGGATTAATAACCAGAATATATAAGGAGCTCAAACAACTCTATAGGAAAAAATCTAATAATCCGATTAAAAAATGGGCAAAAGATCTGAATAGACATTTCTCAAAAGAAGACATACAAATGGCAAACAGGCATATGAAAAGGTGCTCAACATCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
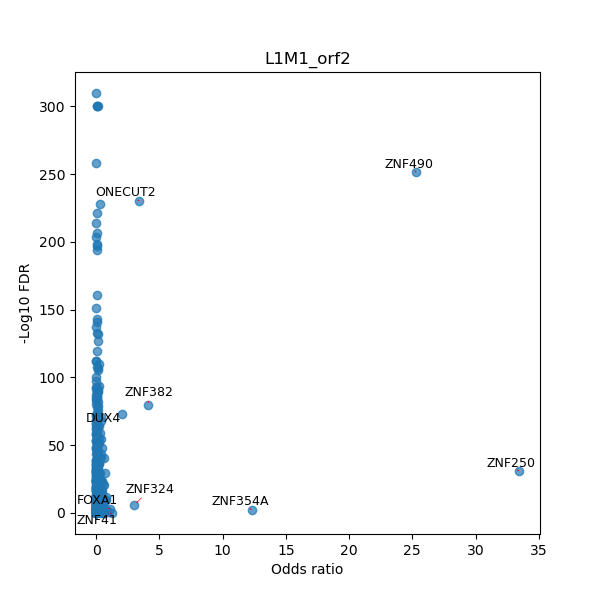
| L1M1_orf2 | ZNF382 | 1191 | 1214 | + | 27.79 | AGGAGACATTACAACTGATACCGC |
| L1M1_orf2 | CDF5 | 2250 | 2270 | - | 17.45 | ATTACTTTTTTGATTTCTTTT |
| L1M1_orf2 | ZNF324 | 1708 | 1721 | + | 17.11 | ATGCAAGGATGGTT |
| L1M1_orf2 | ZNF354A | 293 | 312 | + | 16.84 | TAATGATAAAGGGGTCAATT |
| L1M1_orf2 | OLIG1 | 614 | 623 | - | 16.51 | AACATATGGT |
| L1M1_orf2 | FLC | 2388 | 2401 | + | 16.40 | CAAAAAATGGAAAG |
| L1M1_orf2 | ONECUT3 | 705 | 716 | + | 16.37 | AGAAATCAATAA |
| L1M1_orf2 | PI | 2388 | 2401 | + | 16.35 | CAAAAAATGGAAAG |
| L1M1_orf2 | BHLHE23 | 614 | 623 | - | 16.35 | AACATATGGT |
| L1M1_orf2 | CDF5 | 2042 | 2062 | - | 16.30 | TGCCCTTTATTTCTTTCTCTT |
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.