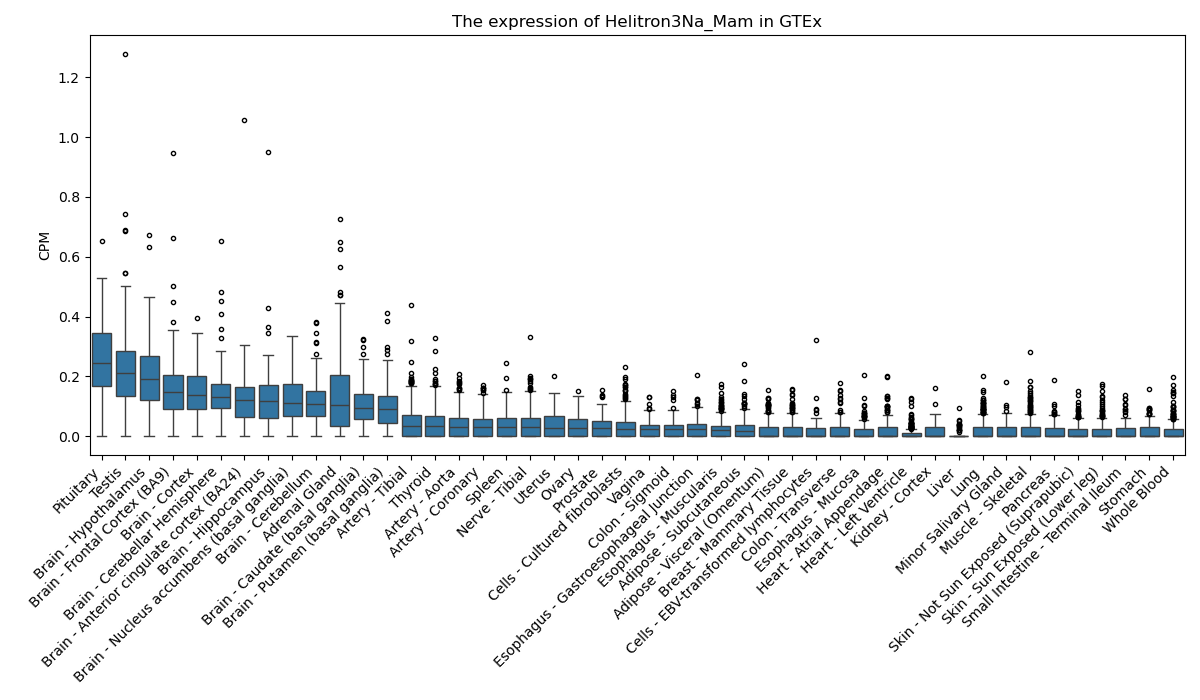
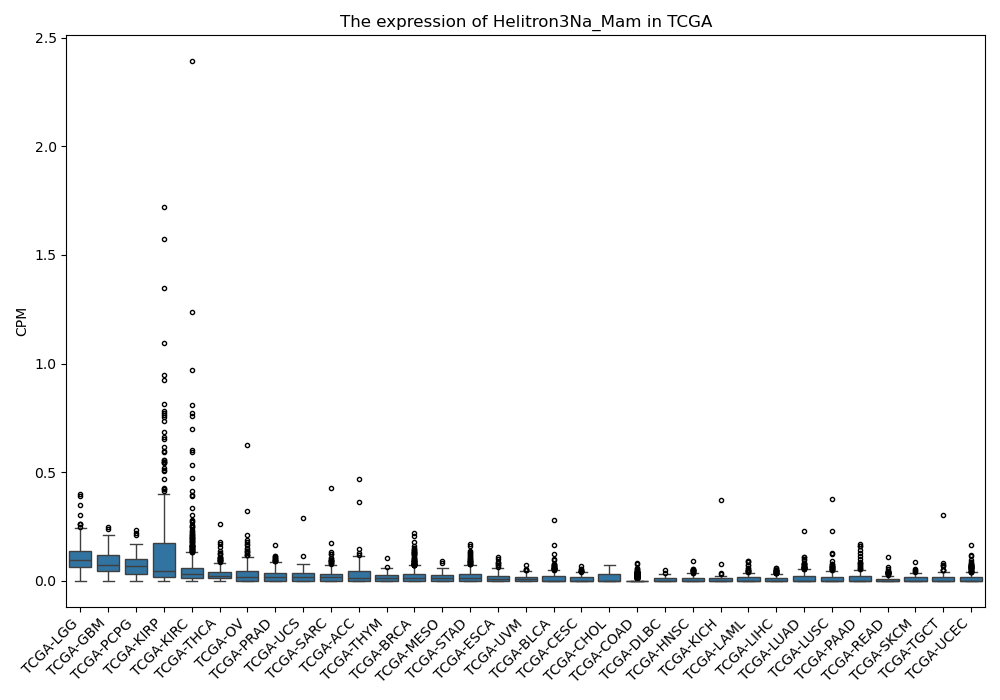
Helitron3Na_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000160 |
|---|---|
| TE superfamily | Helitron-1 |
| TE class | RC |
| Species | Eutheria |
| Length | 486 |
| Kimura value | 31.32 |
| Tau index | 0.8210 |
| Description | Helitron DNA transposon, Helitron3Na_Mam subfamily |
| Comment | Helitrons are thought to transpose with a rolling circle mechanism, and represent a distinct class of DNA transposon. Helitron3Na_Mam represents a non-autonomous helitron fossil also found in other mammals, though it is not clearly similar to the other human helitron models. |
| Sequence |
TCTATATAAATAAAATTAAGTTTCTGTCTGTTTGTCTGTTACCGCATCACGCGAAAATGGCTGGATGGATTTTCACCAAATTTGGAGGGTATGTTTGGGATGGTCTGACTTAAAATATAGGCTATGTGGCATACACGAAATTCACTTTGGGGGGCCCTGGGGGGCACCTCAAAGAGATAGGCAGTCATCCTCCAGAGCAGCTGAAACTGAGGTACAACGAGAGGCCCGTGTGACTGCCGATCGGGAGAGACATGCCATACGTATTAAGACGCCGTGGACAGAGAAAACAAACAGTGGTTTCAAATATGAGCCCCAAATCGAAAGTCACAAGGGCAGACGCTCCGAATTGCAGGCGTTGATTTGCGATCGAGCTGCTTCTCACATGGGCAACTCTATGTAGCGTGCTCCAGGGTGAGTAGCAGCAGGGATTTATTTATTTAACGATGGTTAATGATTCTCCCGAGCAACGCCGGGGAGGCCAGCTAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Helitron3Na_Mam | IDD6 | 29 | 44 | + | 13.05 | TGTTTGTCTGTTACCG |
| Helitron3Na_Mam | TFAP2B | 153 | 165 | - | 13.03 | GCCCCCCAGGGCC |
| Helitron3Na_Mam | TFAP2A | 153 | 165 | - | 13.00 | GCCCCCCAGGGCC |
| Helitron3Na_Mam | Crg-1 | 3 | 13 | + | 12.88 | TATATAAATAA |
| Helitron3Na_Mam | wc-1 | 364 | 370 | + | 12.87 | CGATCGA |
| Helitron3Na_Mam | rib | 50 | 58 | + | 12.77 | CGCGAAAAT |
| Helitron3Na_Mam | sug | 145 | 156 | - | 12.75 | GGCCCCCCAAAG |
| Helitron3Na_Mam | FOXE1 | 430 | 441 | - | 12.74 | TTAAATAAATAA |
| Helitron3Na_Mam | FOXC1 | 3 | 13 | + | 12.70 | TATATAAATAA |
| Helitron3Na_Mam | StBRC1 | 151 | 157 | - | 12.69 | GGGCCCC |
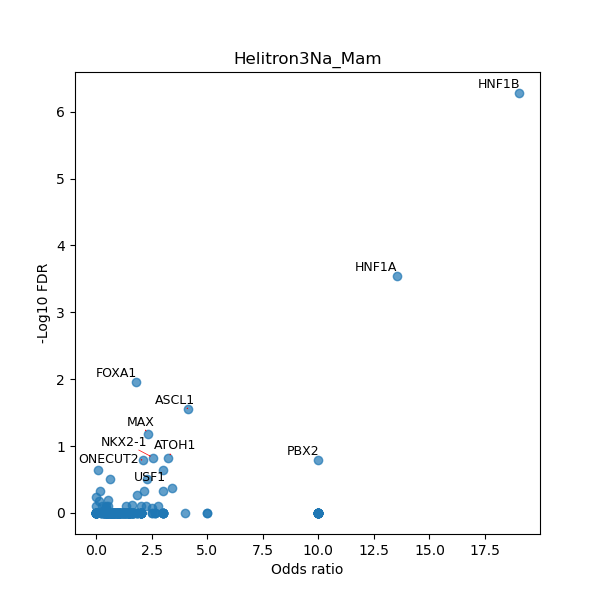
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.