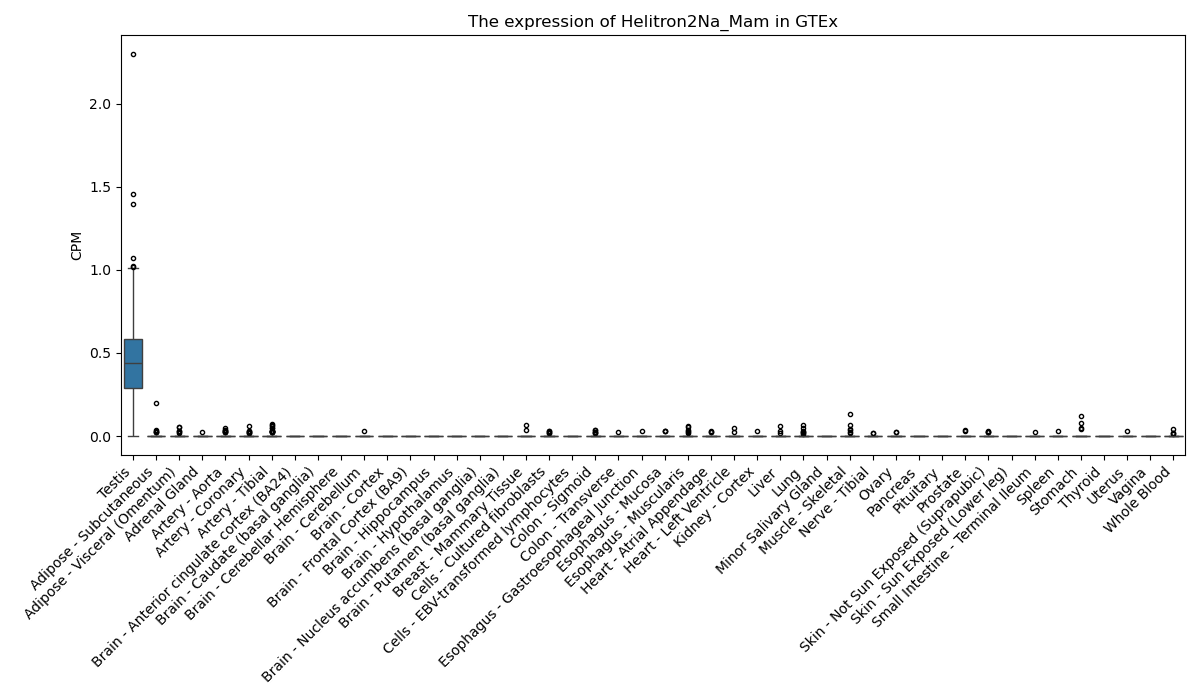
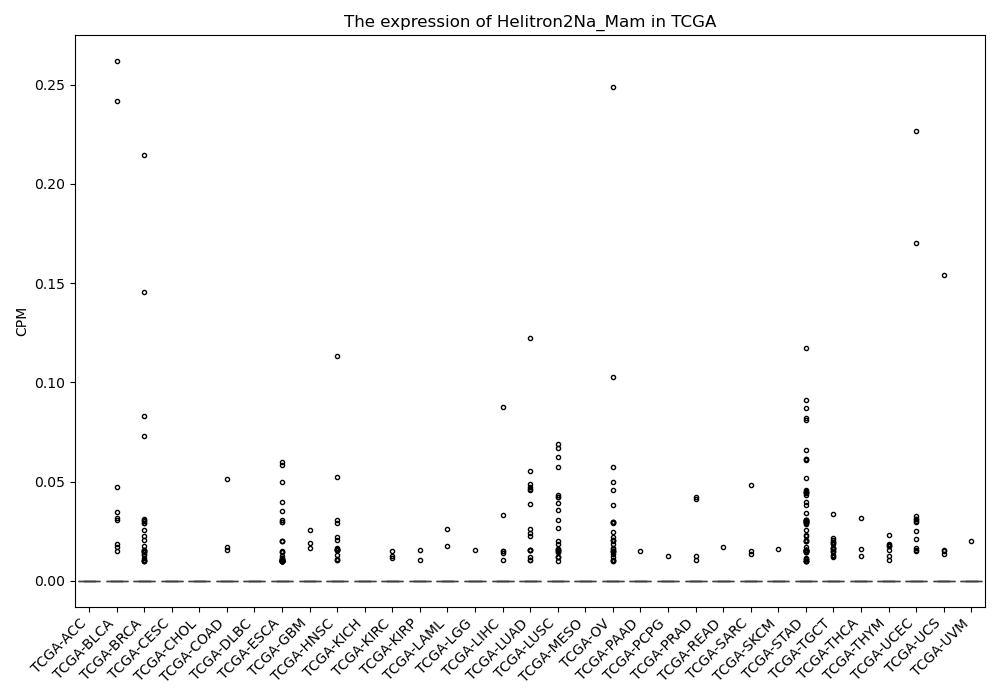
Helitron2Na_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000159 |
|---|---|
| TE superfamily | Helitron-1 |
| TE class | RC |
| Species | Theria_mammals |
| Length | 324 |
| Kimura value | 33.87 |
| Tau index | 1.0000 |
| Description | Helitron DNA transposon, Helitron2Na_Mam subfamily |
| Comment | Helitrons are thought to transpose with a rolling circle mechanism, and represent a distinct class of DNA transposon. While named a helitron, this 324bp TE model its classification is tentative. There is suggestive similarity to a non-autonomous helitron in the sea urchin S. purpuratus. |
| Sequence |
TTGACCGAACGAAGTGAGGTCTCTGTTTCAACTCGACCGCTTGGCATTTTGCATTTGTCTGTCTGTTTGTTCCATGATAACTTTCGAAGGAGTTGACTGATTTTGACCAAATTTGGTACACTACTAAAGGGTATCAAGATACACCTTAAGTTCGATTTGTGGTTCAGTCTGCCATCTTGAAACAAAATGGCAGCCGATAAAAGTTTTCAAATGGGCATATCTTCTGTAATTTTTAATAGATTTCAATGAAATTTGGTACGTGGGTAGAGGTTACAGAGATATGTGATTGCACTGAGTGAAGTCTGCAGTTCACAGAACCACTAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Helitron2Na_Mam | SEP1 | 107 | 119 | + | 13.55 | CCAAATTTGGTAC |
| Helitron2Na_Mam | NFIC | 40 | 46 | + | 13.37 | CTTGGCA |
| Helitron2Na_Mam | ZFP42 | 167 | 179 | - | 13.08 | CAAGATGGCAGAC |
| Helitron2Na_Mam | Hmga1 | 229 | 236 | + | 12.99 | ATTTTTAA |
| Helitron2Na_Mam | WRKY62 | 102 | 109 | - | 12.94 | TGGTCAAA |
| Helitron2Na_Mam | Ddit3::Cebpa | 283 | 292 | - | 12.89 | GTGCAATCAC |
| Helitron2Na_Mam | Yy1 | 172 | 179 | - | 12.89 | CAAGATGG |
| Helitron2Na_Mam | RIM101 | 40 | 46 | - | 12.82 | TGCCAAG |
| Helitron2Na_Mam | AG | 104 | 119 | - | 12.80 | GTACCAAATTTGGTCA |
| Helitron2Na_Mam | Bcl11B | 157 | 165 | - | 12.67 | GAACCACAA |
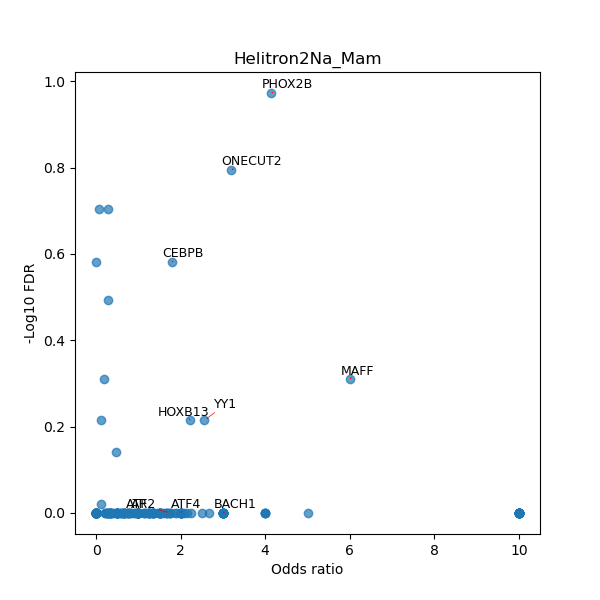
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.