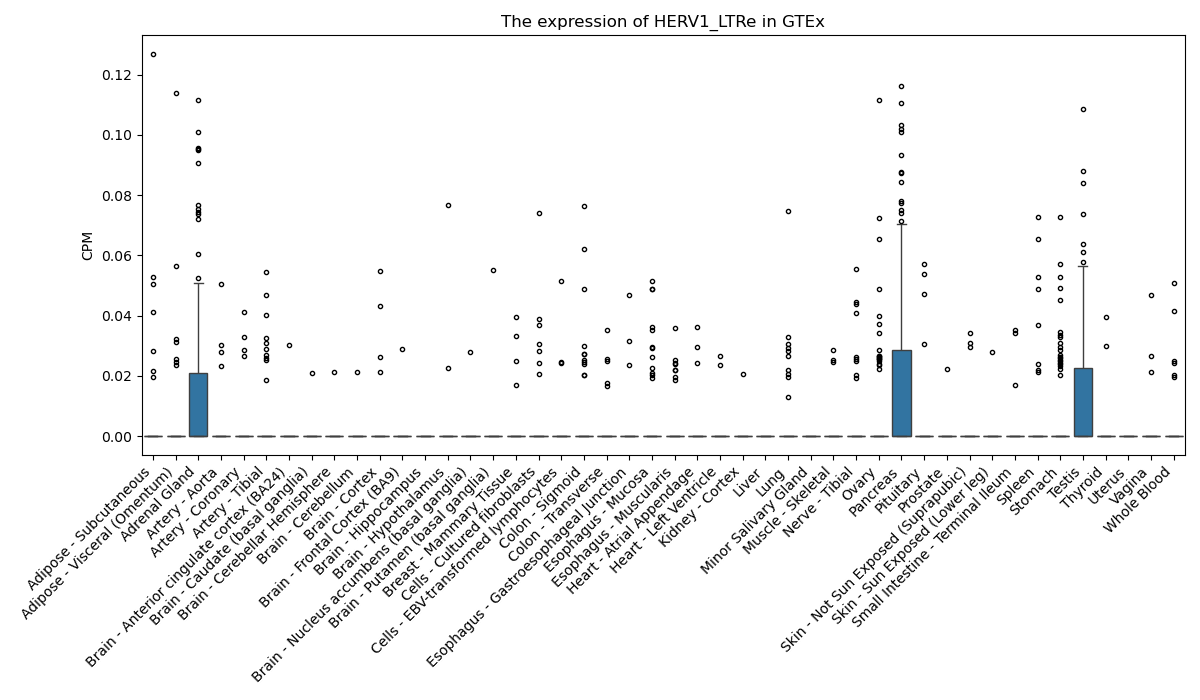
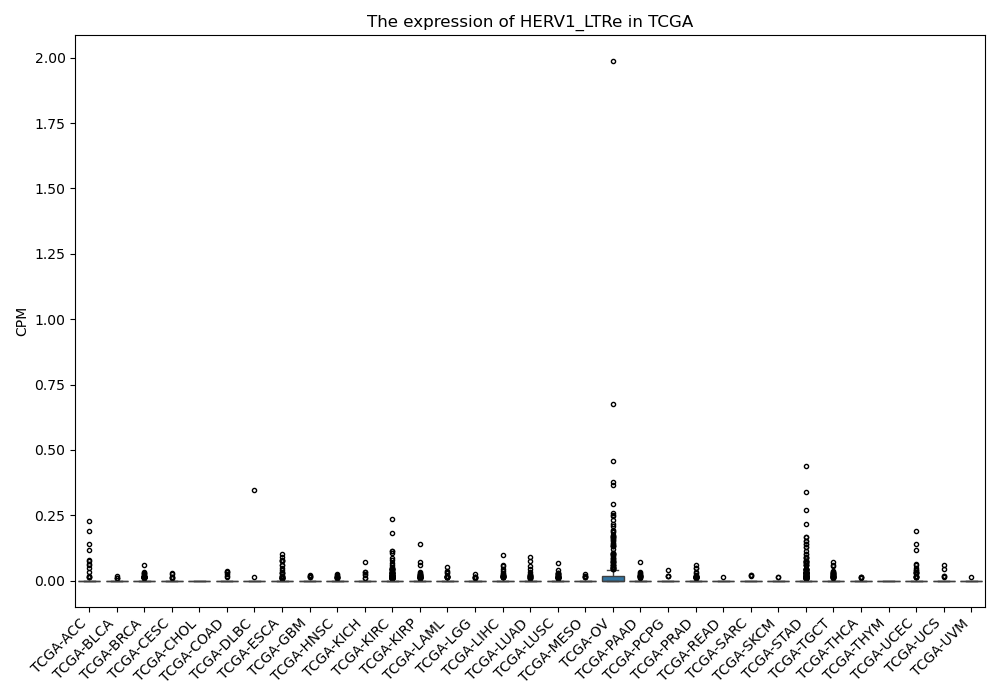
HERV1_LTRe
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000168 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 537 |
| Kimura value | 12.32 |
| Tau index | 0.0000 |
| Description | LTRe (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | HERV1_LTRe represents a mixture of low copy number subfamilies, probably all <10% diverged. |
| Sequence |
TGAAGTAGGAAATTAGGAAAATAACAGAATAATGGCANAAGTAATAATAGCGAAAGTNATAGTAATAGAAANATAATAATAGTTCATAGAATGAATTGCTGTNTTAACCTAGGCTGAAAAGGCTTTAGGTAGCGCACCCCCACCCCCCCCGAAGTNAGAGTTAAGAAAGAATATTAACTGCCTCTCTGTGAAACATTAACCATATCTCTCCCCCGCATATTTTGTAAGTTCTGTAAGTTCCTGTTTTTCTTGCTGTGCAGCTGCAAGGTCACAAGATAGATAAGCGTAAGCTGCAAGACATGTTTTCCCTAAGATGTAAGACTCATGTCACATGATGATTAACTGCTTTTGTTCTCGCTTCTGTAAGCCTGCTTCCTGCNTCACGTANTTCCCGCCTCAAGATGCNTAAAAGGCGCTTGCCTTCTTTGTTCGGTGCTCAGACTTTCAGGACGCATGTCCGCTGAGCCGGTGTACACCTTAAAATAAANCCCTCCTGNACCCCNATCGGTCTCTCCGGTTCCCTGATTTCCCGCTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRe | BRN2 | 282 | 297 | - | 14.47 | CTTGCAGCTTACGCTT |
| HERV1_LTRe | KLF16 | 139 | 149 | + | 14.47 | CCCACCCCCCC |
| HERV1_LTRe | EREB71 | 141 | 150 | + | 14.40 | CACCCCCCCC |
| HERV1_LTRe | MYOG | 257 | 264 | + | 14.35 | GCAGCTGC |
| HERV1_LTRe | MYOG | 257 | 264 | - | 14.35 | GCAGCTGC |
| HERV1_LTRe | IKZF1 | 238 | 245 | - | 14.34 | AACAGGAA |
| HERV1_LTRe | Wt1 | 207 | 216 | + | 14.24 | TCTCCCCCGC |
| HERV1_LTRe | Klf15 | 128 | 149 | + | 14.19 | GGTAGCGCACCCCCACCCCCCC |
| HERV1_LTRe | OSR2 | 357 | 364 | - | 14.17 | ACAGAAGC |
| HERV1_LTRe | MITF | 327 | 336 | - | 14.06 | ATCATGTGAC |
TFBS enrichment in GRCh38
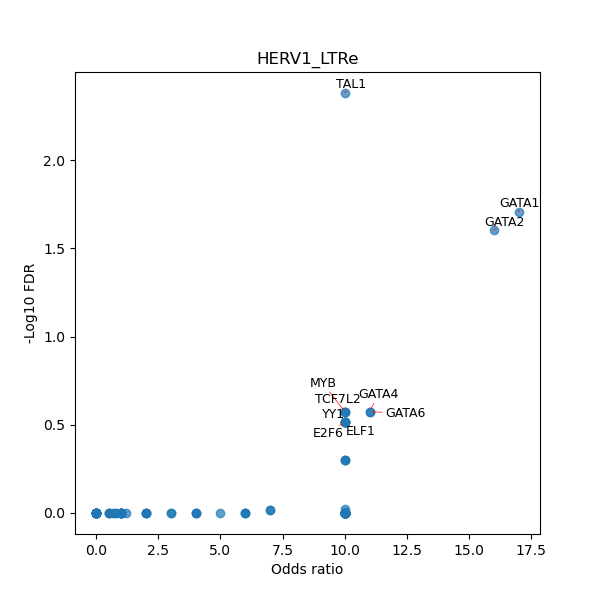
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.