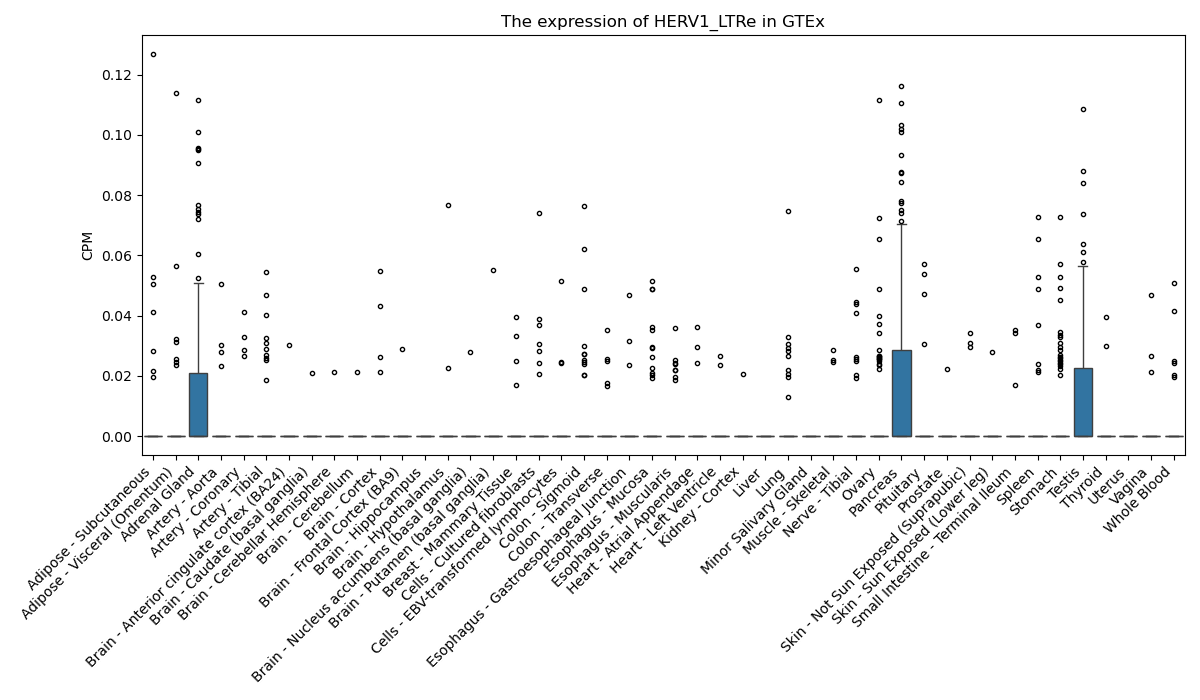
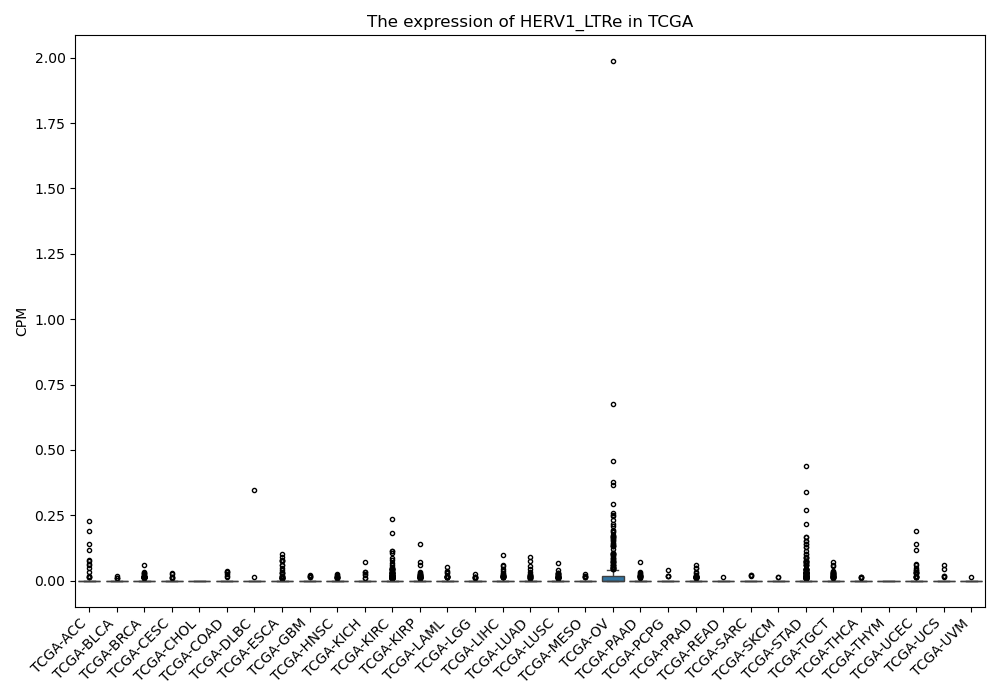
HERV1_LTRe
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000168 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 537 |
| Kimura value | 12.32 |
| Tau index | 0.0000 |
| Description | LTRe (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | HERV1_LTRe represents a mixture of low copy number subfamilies, probably all <10% diverged. |
| Sequence |
TGAAGTAGGAAATTAGGAAAATAACAGAATAATGGCANAAGTAATAATAGCGAAAGTNATAGTAATAGAAANATAATAATAGTTCATAGAATGAATTGCTGTNTTAACCTAGGCTGAAAAGGCTTTAGGTAGCGCACCCCCACCCCCCCCGAAGTNAGAGTTAAGAAAGAATATTAACTGCCTCTCTGTGAAACATTAACCATATCTCTCCCCCGCATATTTTGTAAGTTCTGTAAGTTCCTGTTTTTCTTGCTGTGCAGCTGCAAGGTCACAAGATAGATAAGCGTAAGCTGCAAGACATGTTTTCCCTAAGATGTAAGACTCATGTCACATGATGATTAACTGCTTTTGTTCTCGCTTCTGTAAGCCTGCTTCCTGCNTCACGTANTTCCCGCCTCAAGATGCNTAAAAGGCGCTTGCCTTCTTTGTTCGGTGCTCAGACTTTCAGGACGCATGTCCGCTGAGCCGGTGTACACCTTAAAATAAANCCCTCCTGNACCCCNATCGGTCTCTCCGGTTCCCTGATTTCCCGCTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRe | PATZ1 | 137 | 147 | - | 17.64 | GGGGGTGGGGG |
| HERV1_LTRe | ZNF281 | 138 | 147 | - | 17.16 | GGGGGTGGGG |
| HERV1_LTRe | Klf15 | 132 | 153 | + | 16.86 | GCGCACCCCCACCCCCCCCGAA |
| HERV1_LTRe | Klf6-7-like | 133 | 152 | + | 16.58 | CGCACCCCCACCCCCCCCGA |
| HERV1_LTRe | Klf15 | 134 | 155 | + | 15.94 | GCACCCCCACCCCCCCCGAAGT |
| HERV1_LTRe | KAN2 | 166 | 175 | - | 15.83 | AATATTCTTT |
| HERV1_LTRe | Sox102F | 425 | 436 | + | 15.69 | TTTGTTCGGTGC |
| HERV1_LTRe | FOXO1::ELK3 | 236 | 248 | - | 15.51 | AAAAACAGGAACT |
| HERV1_LTRe | KLF4 | 138 | 145 | + | 15.48 | CCCCACCC |
| HERV1_LTRe | FOXO1::ELK1 | 236 | 248 | - | 15.46 | AAAAACAGGAACT |
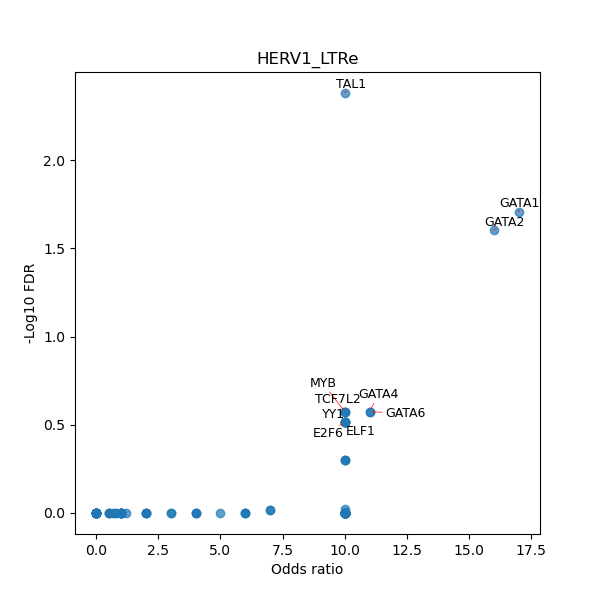
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.