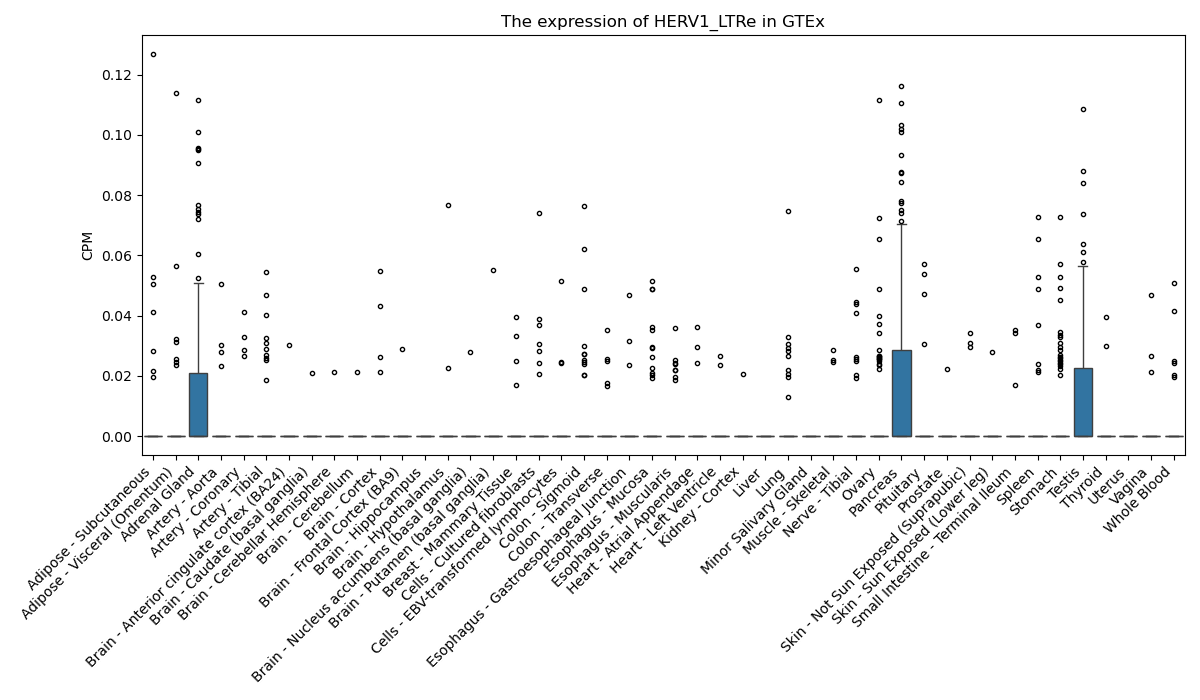
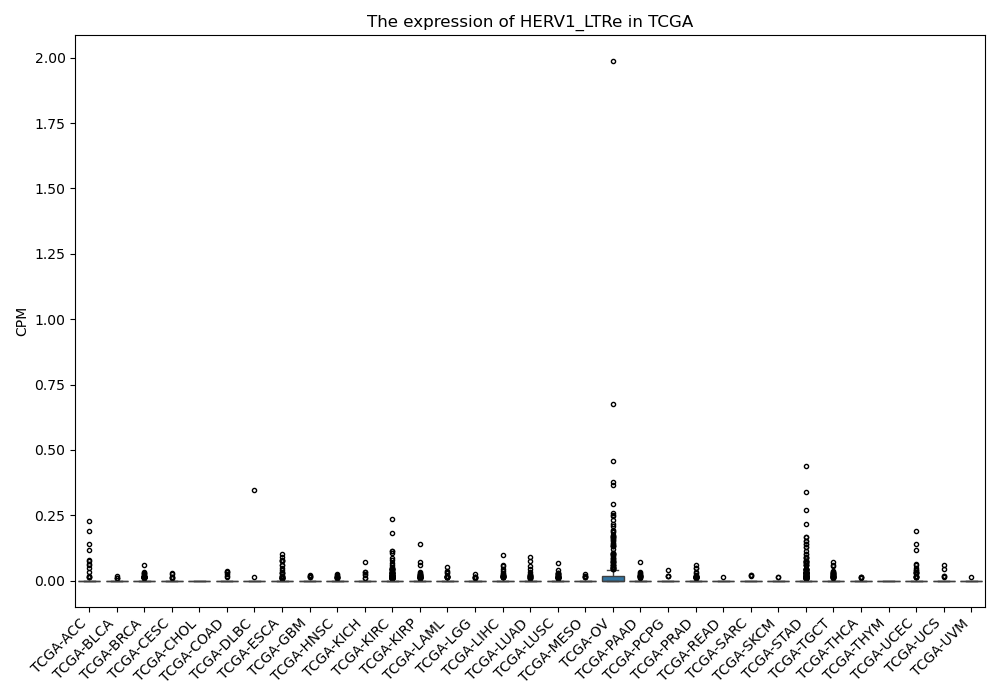
HERV1_LTRe
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000168 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 537 |
| Kimura value | 12.32 |
| Tau index | 0.0000 |
| Description | LTRe (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | HERV1_LTRe represents a mixture of low copy number subfamilies, probably all <10% diverged. |
| Sequence |
TGAAGTAGGAAATTAGGAAAATAACAGAATAATGGCANAAGTAATAATAGCGAAAGTNATAGTAATAGAAANATAATAATAGTTCATAGAATGAATTGCTGTNTTAACCTAGGCTGAAAAGGCTTTAGGTAGCGCACCCCCACCCCCCCCGAAGTNAGAGTTAAGAAAGAATATTAACTGCCTCTCTGTGAAACATTAACCATATCTCTCCCCCGCATATTTTGTAAGTTCTGTAAGTTCCTGTTTTTCTTGCTGTGCAGCTGCAAGGTCACAAGATAGATAAGCGTAAGCTGCAAGACATGTTTTCCCTAAGATGTAAGACTCATGTCACATGATGATTAACTGCTTTTGTTCTCGCTTCTGTAAGCCTGCTTCCTGCNTCACGTANTTCCCGCCTCAAGATGCNTAAAAGGCGCTTGCCTTCTTTGTTCGGTGCTCAGACTTTCAGGACGCATGTCCGCTGAGCCGGTGTACACCTTAAAATAAANCCCTCCTGNACCCCNATCGGTCTCTCCGGTTCCCTGATTTCCCGCTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRe | SP8 | 140 | 150 | + | 15.45 | CCACCCCCCCC |
| HERV1_LTRe | E2F6 | 389 | 396 | - | 15.29 | GGCGGGAA |
| HERV1_LTRe | ZNF768 | 180 | 188 | + | 15.25 | GCCTCTCTG |
| HERV1_LTRe | KLF10 | 138 | 146 | - | 15.20 | GGGGTGGGG |
| HERV1_LTRe | CDC5 | 458 | 466 | - | 15.17 | GCTCAGCGG |
| HERV1_LTRe | GLIS2 | 141 | 154 | + | 15.08 | CACCCCCCCCGAAG |
| HERV1_LTRe | FOXO1::ELF1 | 236 | 248 | - | 15.03 | AAAAACAGGAACT |
| HERV1_LTRe | FOXO1::FLI1 | 236 | 248 | - | 14.91 | AAAAACAGGAACT |
| HERV1_LTRe | USF1 | 327 | 336 | - | 14.82 | ATCATGTGAC |
| HERV1_LTRe | KLF5 | 137 | 146 | + | 14.78 | CCCCCACCCC |
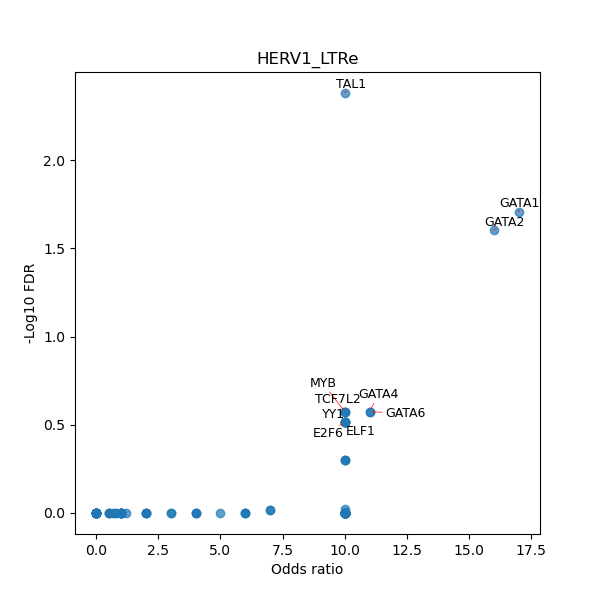
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.