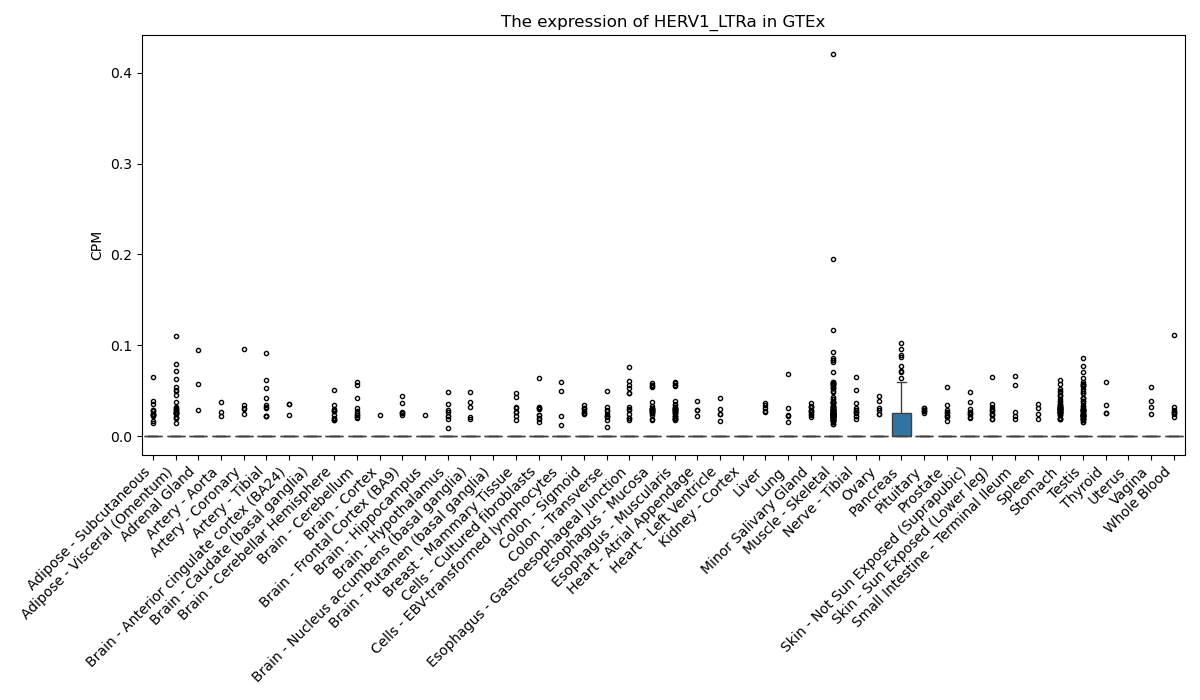
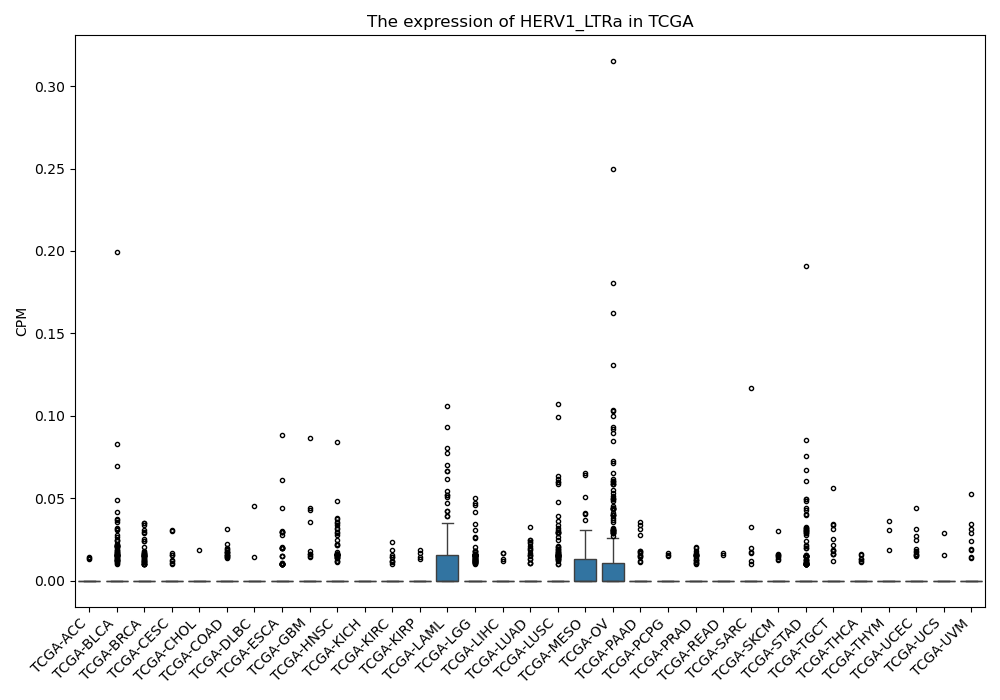
HERV1_LTRa
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000164 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 521 |
| Kimura value | 4.73 |
| Tau index | 0.0000 |
| Description | LTRa (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | - |
| Sequence |
TGAAGAAGGAATTCATGAATTTTACAAGTATAATCAAAGACAACCAAGAAATTTTTACTTTTTCCTTCAAAAGCTAAGTGTAGTGTAGCCCCCACCCGTAGTCTAAGTTAGAGAAGAATACTAACTGCCTGTTTTTCCTTCTGTGCTCAGCGAGCCTTATCTGTACTCGCCAGTTTCACATTCCTTGAGGCTCAGCGAGTTCCTGCTTCACCTCCCTAGCGCAGCTGCAAAGTTACAAGGTTGATACGCGNNCGNCGCAGAAACATGGTTTCCCAGGGATGTGGAACATGTAGTATAGATAAATGTAAAAGACTGATCAACTGCCTTTGTTCTCGCTTCTGTAAGTACGCTTCCTGCATCACGTAGCTCCCGGCCACTGACTGCTTAAAAGGTGGCTGCTTTCTTTGTCCGGGGCTCAGACTTTCCTGGACGCTAGTCCTACTGAGCCAGGTGATCACCTTTTAATAAAGACCTTTCCTGAACTCTGTTCGGTCTCTCCCATCTCTGATTGTCCCGCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRa | Ikzf3 | 348 | 356 | - | 14.00 | CAGGAAGCG |
| HERV1_LTRa | ZKSCAN5 | 207 | 215 | - | 13.73 | GGAGGTGAA |
| HERV1_LTRa | DOF4.7 | 55 | 63 | + | 13.72 | TTACTTTTT |
| HERV1_LTRa | ASCL1 | 221 | 228 | + | 13.67 | GCAGCTGC |
| HERV1_LTRa | ASCL1 | 221 | 228 | - | 13.67 | GCAGCTGC |
| HERV1_LTRa | PATZ1 | 89 | 99 | - | 13.65 | ACGGGTGGGGG |
| HERV1_LTRa | NAC68 | 24 | 31 | + | 13.51 | ACAAGTAT |
| HERV1_LTRa | KLF7 | 90 | 97 | - | 13.19 | GGGTGGGG |
| HERV1_LTRa | Sox7 | 324 | 333 | - | 13.18 | AGAACAAAGG |
| HERV1_LTRa | SOX4 | 324 | 331 | - | 13.17 | AACAAAGG |
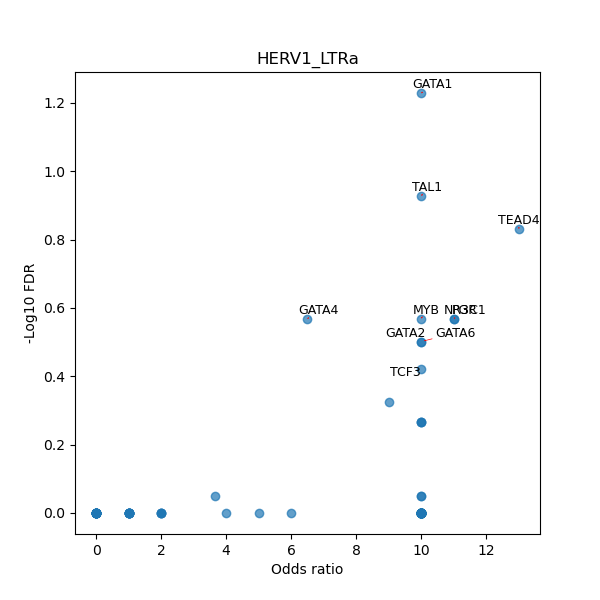
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.