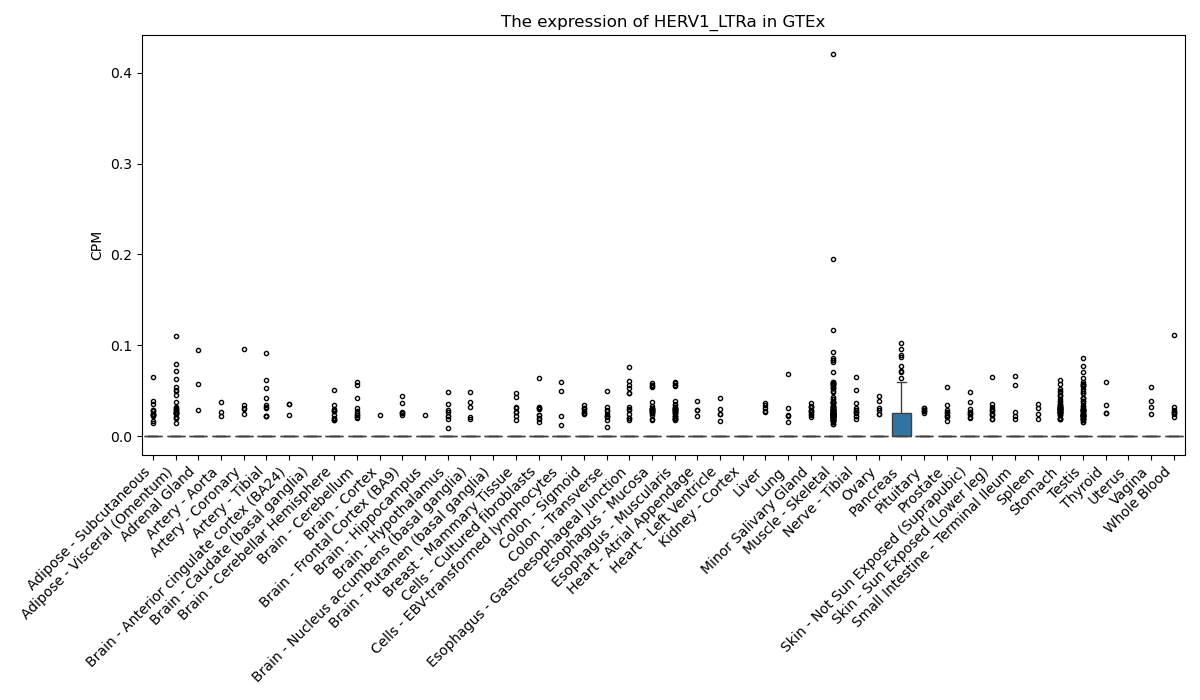
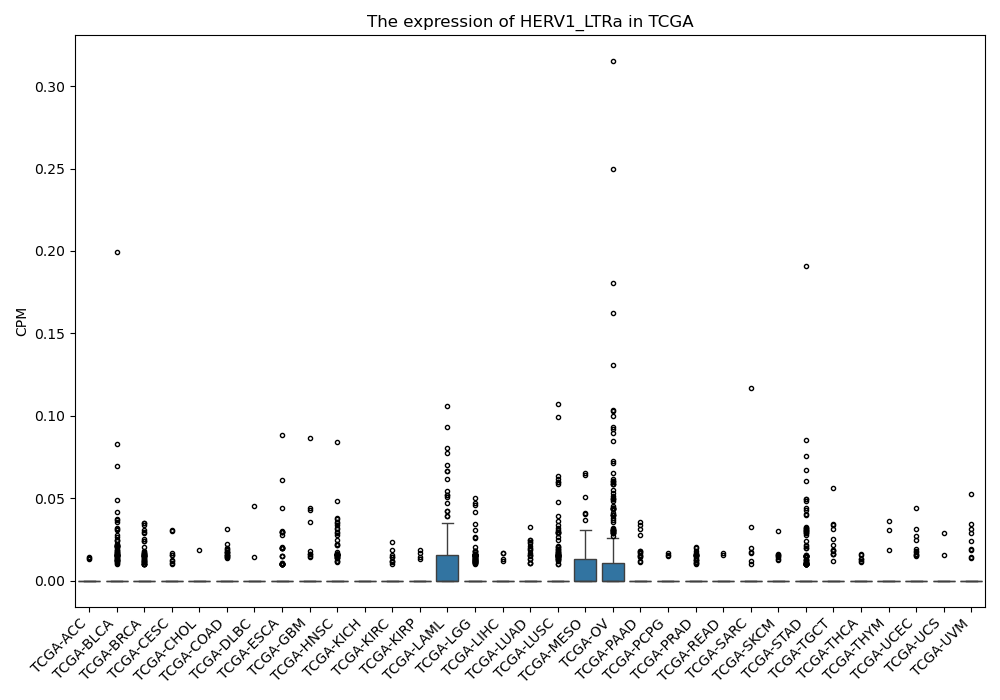
HERV1_LTRa
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000164 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 521 |
| Kimura value | 4.73 |
| Tau index | 0.0000 |
| Description | LTRa (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | - |
| Sequence |
TGAAGAAGGAATTCATGAATTTTACAAGTATAATCAAAGACAACCAAGAAATTTTTACTTTTTCCTTCAAAAGCTAAGTGTAGTGTAGCCCCCACCCGTAGTCTAAGTTAGAGAAGAATACTAACTGCCTGTTTTTCCTTCTGTGCTCAGCGAGCCTTATCTGTACTCGCCAGTTTCACATTCCTTGAGGCTCAGCGAGTTCCTGCTTCACCTCCCTAGCGCAGCTGCAAAGTTACAAGGTTGATACGCGNNCGNCGCAGAAACATGGTTTCCCAGGGATGTGGAACATGTAGTATAGATAAATGTAAAAGACTGATCAACTGCCTTTGTTCTCGCTTCTGTAAGTACGCTTCCTGCATCACGTAGCTCCCGGCCACTGACTGCTTAAAAGGTGGCTGCTTTCTTTGTCCGGGGCTCAGACTTTCCTGGACGCTAGTCCTACTGAGCCAGGTGATCACCTTTTAATAAAGACCTTTCCTGAACTCTGTTCGGTCTCTCCCATCTCTGATTGTCCCGCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRa | DOF1.5 | 53 | 68 | - | 18.09 | GAAGGAAAAAGTAAAA |
| HERV1_LTRa | NHLH1 | 220 | 228 | + | 16.93 | CGCAGCTGC |
| HERV1_LTRa | GATA1::TAL1 | 157 | 173 | + | 15.95 | TTATCTGTACTCGCCAG |
| HERV1_LTRa | DOF1.7 | 53 | 63 | - | 15.76 | AAAAAGTAAAA |
| HERV1_LTRa | KLF4 | 90 | 97 | + | 15.48 | CCCCACCC |
| HERV1_LTRa | FOXO1::ELF1 | 124 | 136 | - | 14.77 | AAAAACAGGCAGT |
| HERV1_LTRa | KLF1 | 90 | 97 | - | 14.64 | GGGTGGGG |
| HERV1_LTRa | D | 323 | 333 | + | 14.54 | GCCTTTGTTCT |
| HERV1_LTRa | Sox11 | 325 | 332 | - | 14.51 | GAACAAAG |
| HERV1_LTRa | ERF::FOXO1 | 125 | 136 | - | 14.49 | AAAAACAGGCAG |
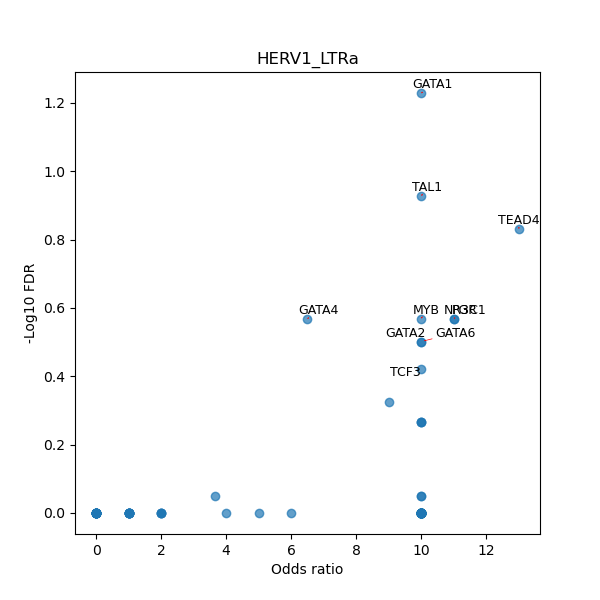
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.