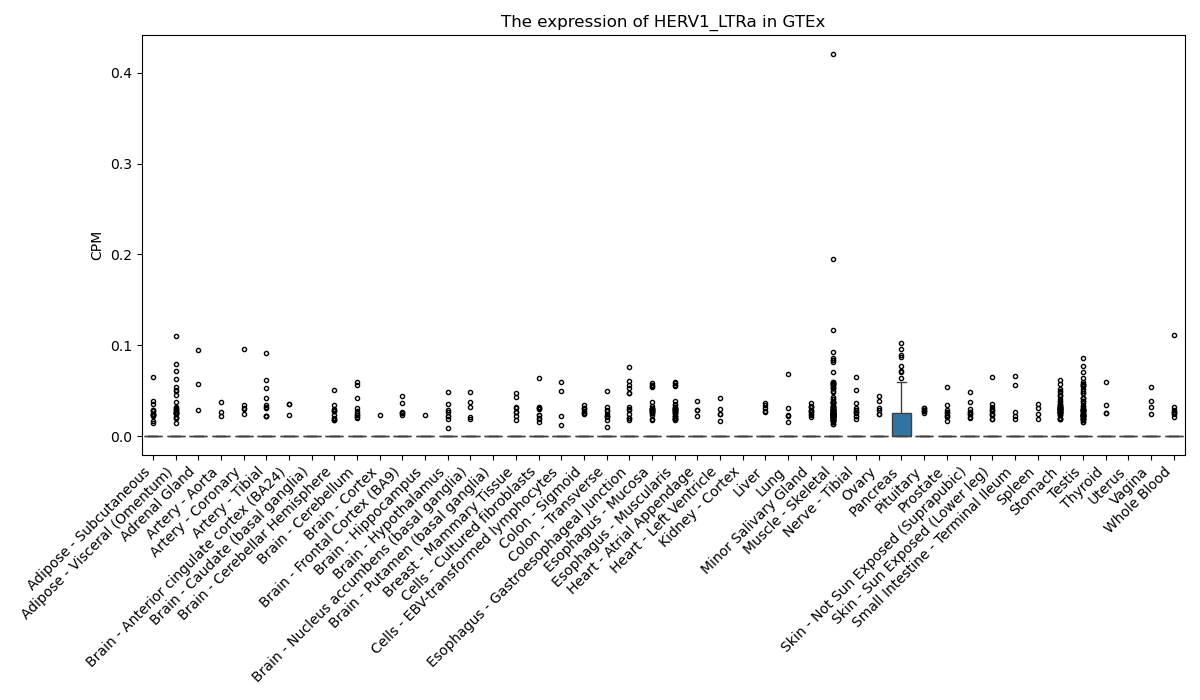
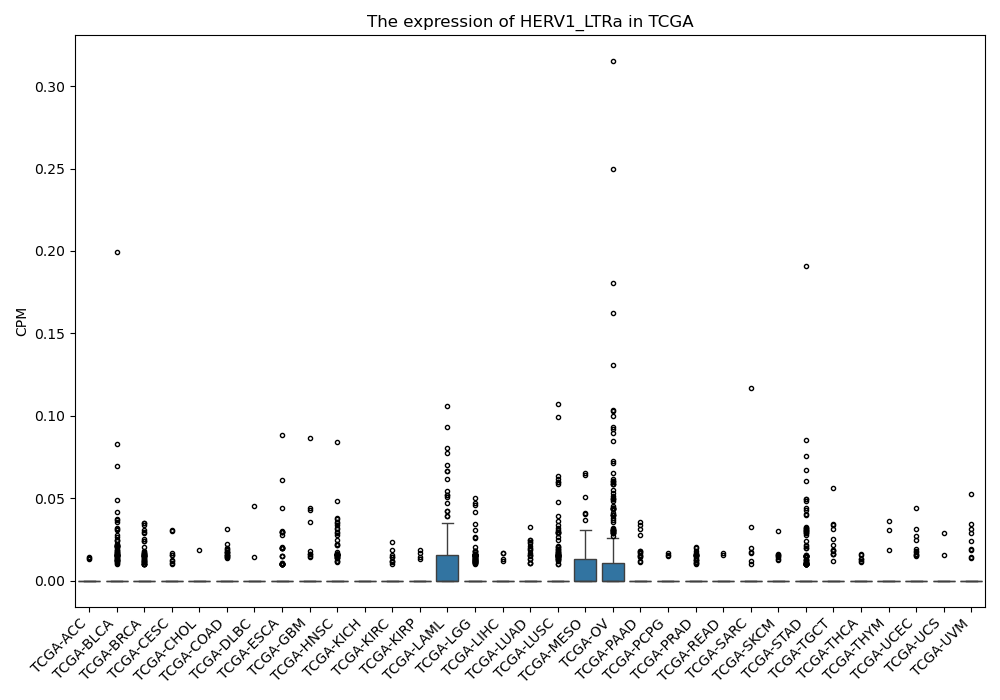
HERV1_LTRa
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000164 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 521 |
| Kimura value | 4.73 |
| Tau index | 0.0000 |
| Description | LTRa (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | - |
| Sequence |
TGAAGAAGGAATTCATGAATTTTACAAGTATAATCAAAGACAACCAAGAAATTTTTACTTTTTCCTTCAAAAGCTAAGTGTAGTGTAGCCCCCACCCGTAGTCTAAGTTAGAGAAGAATACTAACTGCCTGTTTTTCCTTCTGTGCTCAGCGAGCCTTATCTGTACTCGCCAGTTTCACATTCCTTGAGGCTCAGCGAGTTCCTGCTTCACCTCCCTAGCGCAGCTGCAAAGTTACAAGGTTGATACGCGNNCGNCGCAGAAACATGGTTTCCCAGGGATGTGGAACATGTAGTATAGATAAATGTAAAAGACTGATCAACTGCCTTTGTTCTCGCTTCTGTAAGTACGCTTCCTGCATCACGTAGCTCCCGGCCACTGACTGCTTAAAAGGTGGCTGCTTTCTTTGTCCGGGGCTCAGACTTTCCTGGACGCTAGTCCTACTGAGCCAGGTGATCACCTTTTAATAAAGACCTTTCCTGAACTCTGTTCGGTCTCTCCCATCTCTGATTGTCCCGCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRa | ac | 221 | 228 | + | 14.47 | GCAGCTGC |
| HERV1_LTRa | ac | 221 | 228 | - | 14.47 | GCAGCTGC |
| HERV1_LTRa | MYOG | 221 | 228 | + | 14.35 | GCAGCTGC |
| HERV1_LTRa | MYOG | 221 | 228 | - | 14.35 | GCAGCTGC |
| HERV1_LTRa | Sox6 | 324 | 333 | + | 14.21 | CCTTTGTTCT |
| HERV1_LTRa | GATA4 | 155 | 162 | + | 14.20 | CCTTATCT |
| HERV1_LTRa | OSR2 | 335 | 342 | - | 14.17 | ACAGAAGC |
| HERV1_LTRa | FOXO1::ELK1 | 124 | 136 | - | 14.11 | AAAAACAGGCAGT |
| HERV1_LTRa | Thap11 | 281 | 294 | - | 14.06 | ACTACATGTTCCAC |
| HERV1_LTRa | DOF3.2 | 55 | 70 | + | 14.03 | TTACTTTTTCCTTCAA |
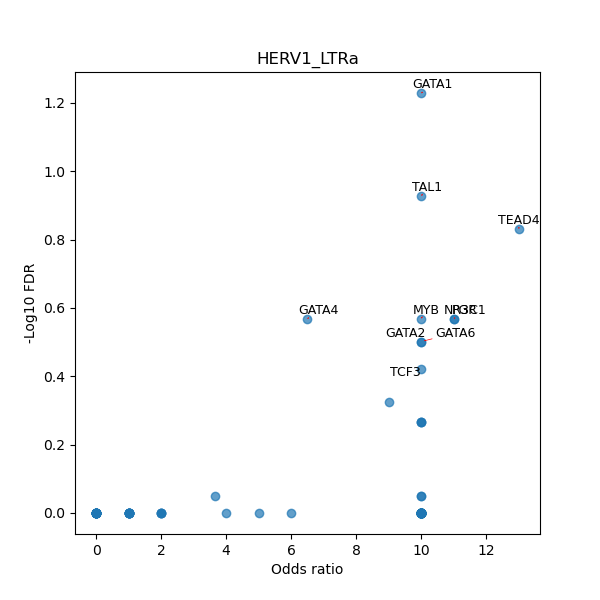
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.