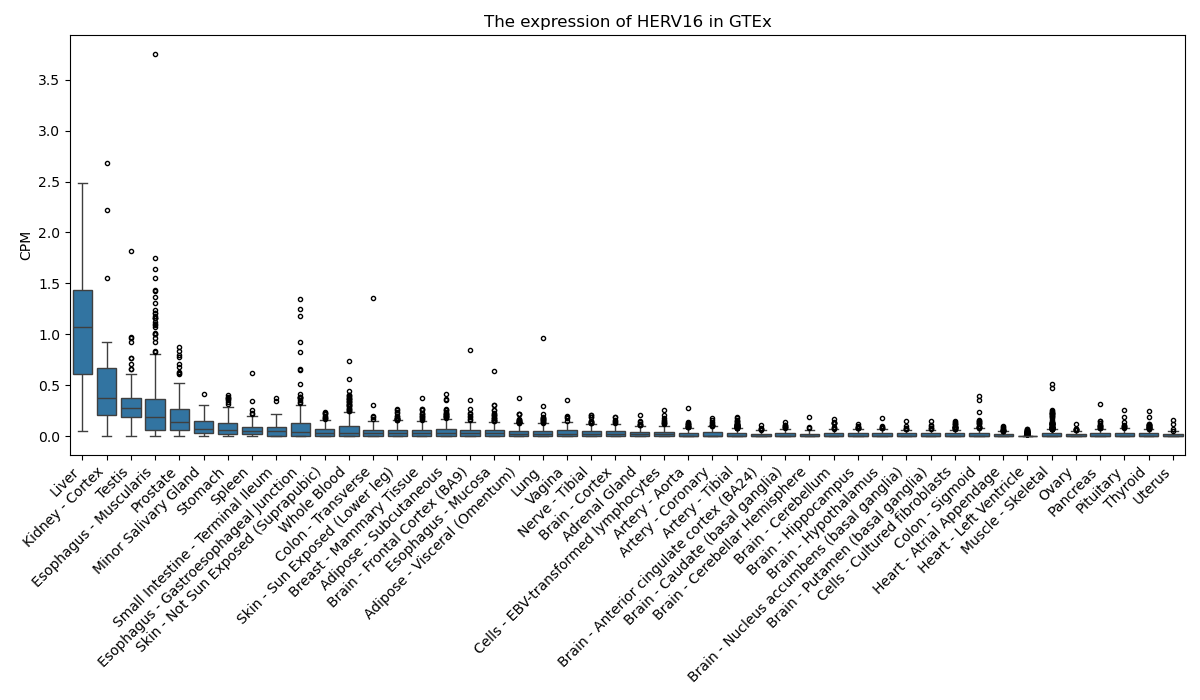
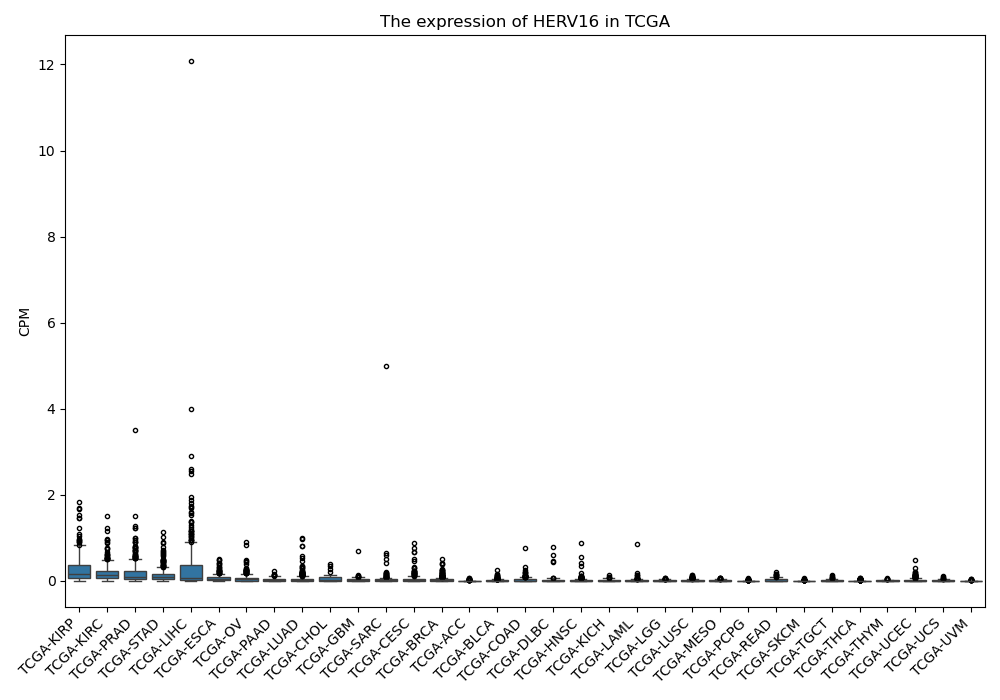
HERV16
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000162 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Theria_mammals |
| Length | 4996 |
| Kimura value | 32.10 |
| Tau index | 0.9660 |
| Description | ERVL endogenous retrovirus, HERV16 subfamily (internal) |
| Comment | LTRs of HERV16 are listed in Repbase as LTR16 sequences. HERV16 is related to HERVL and is relatively old (>20% divergence from consensus). Bases 300 to 1300 encoded a GAG protein closely similar to the murine "retrovirus restriction polypeptide" (PIDs e24674 and e242676). |
| Sequence |
GTTGGTACCAGGAGTGGTCCGAGAAAGCAGACGNTNCTAAGATGGGATTTTGGAGCTGGATCACCCGCCGNCCGGCTGGCAATGAGGACCCCATCACTGGTGGTAGGTGGAGCACGGATAGCCCCTGGCACGAGGTAGCGGTGCAATTGTTAAAACTTTCACCGGTGGTGAACTGGGATGGNATACCGGTGGAAAGAGAATGCACTGGCNGGTGCAATGTNTCAGGCGTTTGAGAAATATGGGANGAATTAANTACATGNAAGGACAATGGAATTGGATGGCTGTTGCTAAGCNCGACTGACGCTCTGGAAAAAGACAATGAAAGGCTGAGAGCGATTAATCGNCAATTNAAAGCTAAGTGTGAAAGCCAGAGGGCCTCCTTGGCAGCATATAAAGAGACTCTCATCTCCTGCAGCCGGAGGGCAGAGAAAGCTGAGGATCAGGCCCAGGACTTAATCAGAGTAGCAGAGCTCCAGAGAAGGTTGAATTCTCAACCNAGGCAGGTCTGCTATGCCAAGGTCAGGGCCCTGGTTGGGAAAGAATGGGACCCTGANACNTGGGATGGGGACATCTGGGTCGATGCNCCTGAAAATCTTGAATCCCCAGATTCCCCTGAACCCTCTGGGCCTGCAGAAGTGGCCCACTCCTCCCTGTTAAGGGCTAGCACTCNCTCCTTGCTTCGCGNGAAGACGATGCAGAGGCCTCTNCCTTNGCAAGACAACATGCGCCCCCCTCAGGATCTGCCCCCACCTCCCCTCCTGGCCACCAGACCAATAACTAGGGTTAAGTCACAGCATAACCCGGCCGGGGAAGTGCTGGGCCTGNTAAGGGAGGAAAGGGACTATACCCCAAAGGAGCTGCAGGACNCGNCTAGCCAGCATGTACCGGCAGGACCGGGAGAGTACGCATGGGACTGGATTCTGAGGGTGCTGGATCAAGGGGGNCGGAACATAAAGTTGGATAAGGGAGAGTTTATCGATNTGGGAGCACTCTCCCGNGATACAGGATTTAACACCCTGGCAAGGACCCCGGGAGATGGTGCNAACACGCTGCTAGGATGGCTCCTGGAAGCNTGGAGAAAGCGATGGCCCACACTAAGTGAAGTAGAAATGCCAGAACTGCCGTGGCAGACGGTGGAAGAAGGGATCAAAAGGCTCAGAGAAGTGGGCATGCTAGAGTGGATATACTATGTAAGGCCGGAAAACCCACCAGATGACTATGTTCCGCGGGAGGGCCCAGAGGACACNCCATTTACCAAAGCNATAAGGAATGCGCTGGTGAGAGGGGCACCAGCATCACTGAGAAGCTCAGTGGTGGCTNTCCTCTGCAGGCCAGGGCTGACGGTAGGAGANGCCGTTACAGAACTGGGCTCNCTGATAGCAATGGGGATGATAGGACCCCGAAATAATAGAGGCCAGGTGGCGGCGCTTAACCGTCAGAAGCAAGGTGGGCGCAATTATCGTAATGNACGGCAAGGTCGGAGTGGCAGCCGGGGGGGCCTGACCCGCAGAGAGCTATGGAGATGGTTAATAGAACACGGCGTCCCTAGGGGCAAGATAGATGGGCAGCCAACAGGAAGNCATGGACAGTTAATAGAACATGGCGTCCTAGGGGCAAGATAGATGGGCAGCCAACAAGGGTGTATTGCTCAACTNAAACAANCAAAAGAAATCAAGGATGGATGANCAGGAGGCTGAGGGCAGTCGCCCCAATAAAAAGTCACGATCCCTTGCCCAGTTTCCGGACCTGAGCCAGTTTTCAGACCCGGAACCCATTGACTGAAGGAGAGGCCGGGTCCCCAGGAGGAAGGACCCTGCAACACCACGGCAAGTGTACACGGTAATGATTCCCCCAGTCCTTCCCCAAAGGGACCTACGGCCATTTACTCTGGGTAACCGTACACTGGGGAAAGGGGAATACCCAGACATTTCGAGGACTGTTGGACACAGGGTCCGAGTTGACATTGATACCCGGAGACCCGAAGCGTCATCATGGCCCTCCCGTTAGAGTGGGGGCATATGGGGGCCAGGTAATAAATGGAGTCCTGGCCCAGGTCCGGCTCACAGTGGGTCCACTGGGTCCACGGACCCACAGTGGTCATTTCCCCGGTCCCCGAATGTATAATTGGAATGGACATACTTGGTAGTTGGCANAACCCCCACATTGGTTCCTTGGCCTGTGGGGTAAGAGCTATCATAGTGGGGAAGGCCAAGTGGAAGCCTCTGAAACTGCCCTCACCTTCTCCGGNCAAGATAGTAAATCAAAAACAATATCGCATCCTCGGGGTGGAGAATGGCAGAGATTAGTGCCACCCTTAAAGACCTAAAGGATGCAGGAGNTGTGGTCCCCATCATATCTCCATTTAATTCACCAGTNCNGGCCCCTGCAAAAACCAGACGGATCCTGGANGACGACAGTGGACTACCGCAAACTCAACCAAGTAGTANCCCAATTGCAGCTGCTGTGCCAGATGTGGTATCTTTGCTAGAGCAGATTAACACGGCCTCAGGTACGTGGTATGCGGCCATTGATCTGGCGAATGCGTTCTTTTCCATCCCTATCAGAAAGGAGGATCAGAAGCAGTTCGCATTCACNTGGAACGGACAACAGTATACATTTACAGTCTTGCCCCAGGGCTNTGTTAACTCTCCTGCCCTCTGTCATAATATAGTCCGAAGGGACCTGGACCATCTGGACATTCCGCAGAACATCACATTGGTCCACTATATCGATGACATCATGCTAATCGGACCGGAATGAGCAAGAAGTGGCAAGTACGTTGGAGGCCTTGGTAAGACACATGCGCTCCAGAGGGTGGGAGATAAACCCTACGAAGATTCAGGGGCCTGCCACATCAGTGAAGTTTTTAGGGGTCCAGTGGTCTGGGGCATGCCGGGACATCCCCCCAAAGTAAAGGACAAATTATTGCATCTTGCACCTCCCACCACNAAGAAGGAAGCACAACGCCTGGTAGGCCTCTTCGGGTTCTGGAGGCAGCATATTCCACACTTGGGAATACTGCTCCGACCCATNTACCGGGTGACGCGAAAGGCTGCCAGCTTTGAGTGGGGCCCAAGCAGGAAAGGGCTCTGTAGCTNACCGCATATCGTGTNCGAANCGCANANNCGGNCCTCCAAGTCATAAAGTNGGGCGGGCCCAGCAGCAGTCCATCATAAGGTGGAAATAGNATATCCGAGATTAAGCCCGAGNAGGACCAGAGGGCGCAGGTNAGNTNCATGAGCAGGTAGCCCAGACCCCCATGNCACCCACCACNGTTGCACCAGCGCCTCTCCTTCGGCTCGCACCTATGGCCATATNGAGGNNTGTGGTCCCGTATGACCAGCTGAAGGAAAGCTCGAGCTTGGTTTACAGGATCAGCTCGGTATGTGGGCGCAAGCCAAAATANGTGGTGGCTGCACTACAGCCCCATTCAGGGGTGGCCCTGAAAGGCAGCAGGGAGTGAAAATATCTTCCCAGTGGGCAGAGCTGCGAGCAGTGCACCTGGTCATCCACTTTNCGTGGAAGGAGAAGTGGCCCGAGATGGTAATATATACGGATTNCTGGACAGTGGCCAATGGCCTGGTCGGCTGGTCAGGAGCTTGGAAGGAAAAGATTAGAGANAGGGAAGTCTGGGGTAGAGGCATGTGGATAGACATATNGGAAGTGGCACGAAATACGAAGATTTTTGTATCGNTNNTATGTTAACGCTCACANGAAAGCATCNACCATGAAAGAGNCACTGAACAACCAAGTAGACAAAGTGACTTCGACCACTGACGTTAGCCAGCCTTCGTCCCGGCCTCCTCAGANCTGGTNCAACGGACATCTCAANAAAGTAGCCATGGTGGCAGAGATGGAGGCTACGCATGGGCCCAACAGCATGGACTCCTATNCACCAAGNCGATCTAGCTGCTGCCCCGTNTGAATGTCCAACCTGCAGCAACAGAGACCAATGCTCTACCTCNAATACGGCACTATTTATGGAGACCACTTAGCGGCGAATTGACTATATTGGACGCCTTCCATCCTGGAANGGNCAGNGATTCATTCTCACAGGAATANATACNTATTCCGGGTATGGGTTTGCCTTTCCTGCCTGCANAGCCTCATCNAGCATCTGAGGGNCTTAGGAGTGCCTGATCCACAGGCATGGAATCCCACACAACATAGCATCTGANCAGGGCGCTCACTTCACAGCAAAGAAGNGGCACAAGTGGGTCCANAATAACGGGATCCACTTATCNTATCACGTACCGCACCATCCAGAAGNAGCTGACCTNATAGAACGCTTCTGAAGGCACAGNTGAAGCACCAGTNGAGGCGAACTCTGAAAGAATTGGGTGCCATCCTCCAGGATGCAGTATATACATTGAATCANAGGCCTATNCGTNGGGCNTTGNNTCTAATNGGGAGAATGGGATTAGGAATAAGAGGTGAAAGCAGGAGTGTGCCTACTCCCCATCGTAGTGGCCCACAAGGATCACAGCAGTGGTTTGAACACGGATTGGACTACTGATATCCAGGAATCGCCGCACAAGAAGGNGAATCCACATTTGGCAGGAGTAATTGACCNTGATCANCAGGAGGGGTAGGGCTGCTNTCGCANAATGAGGAAGAAAGGAATATGTNTGGAACCAGGTGATCCNCTTGGGTGCCTCCTGGTACTCCCTTGCCCCATTTAACTGTAAATGGACAATNCCACCGAAAAGAGCTCGGACCNCTCCGGAATGAAGGTTTGGNTCACATCGCCNGGTAAGCCACCAAGACCTGTCGAAGTGATAGCTGNGTGAGGGANATCTAGAATGGATAGTAGAAGAGGGAGACGATGAGTACCAGTTNCGGCCCCGAGACCAACTGCAGCAANNGGGGCTGTAGTTCGTCCCACTAACCTCCCTCTTCTAAGTTTCGCTTCAGAAAGAGAAGCTTAAAGGAATAATGGAGGAACTGCTCCNNGAACCTGNGTGGAGAAGTAGATCCGTCGAGTACAAAGGTGGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV16 | ZNF157 | 2601 | 2621 | + | 20.85 | CGGACAACAGTATACATTTAC |
| HERV16 | RAX3 | 99 | 110 | + | 20.72 | GGTGGTAGGTGG |
| HERV16 | MYB63 | 100 | 110 | - | 18.49 | CCACCTACCAC |
| HERV16 | TCP13 | 2341 | 2351 | - | 18.14 | ATGGGGACCAC |
| HERV16 | PLAG1 | 2011 | 2024 | + | 18.04 | GGGGGCATATGGGG |
| HERV16 | ZNF707 | 5 | 19 | - | 17.96 | GACCACTCCTGGTAC |
| HERV16 | TCP17 | 2341 | 2351 | + | 17.95 | GTGGTCCCCAT |
| HERV16 | ZmbZIP57 | 2732 | 2741 | - | 17.44 | ATGATGTCAT |
| HERV16 | ZmbZIP25 | 2731 | 2740 | + | 17.39 | GATGACATCA |
| HERV16 | ZmbZIP72 | 2731 | 2740 | + | 17.37 | GATGACATCA |
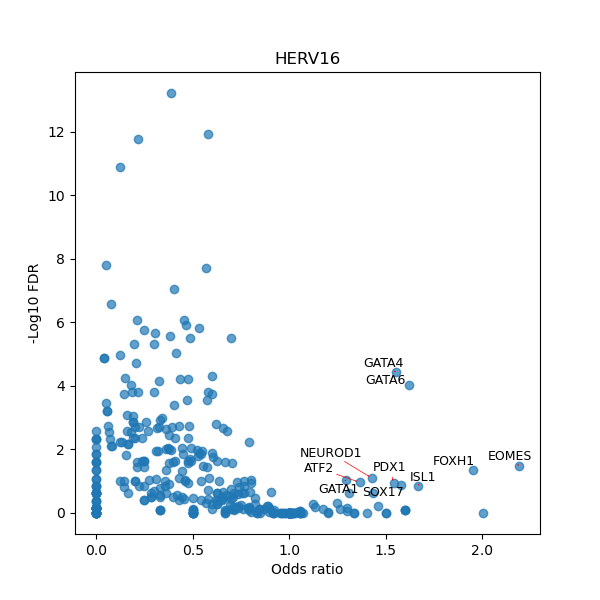
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.