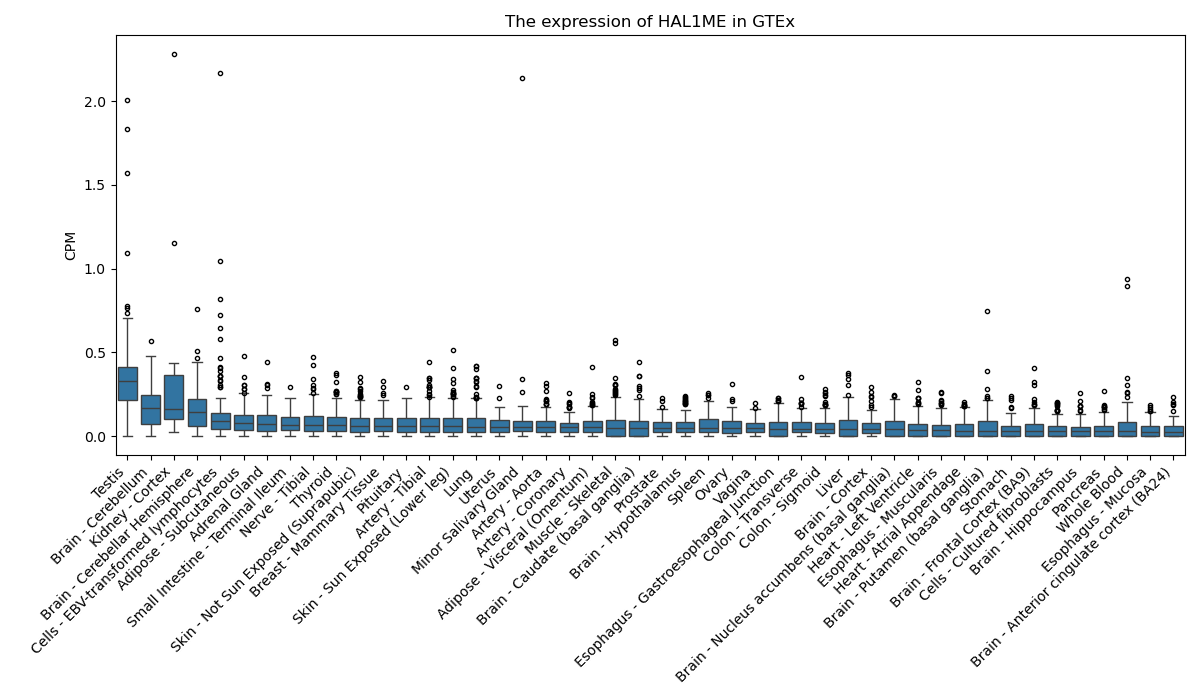
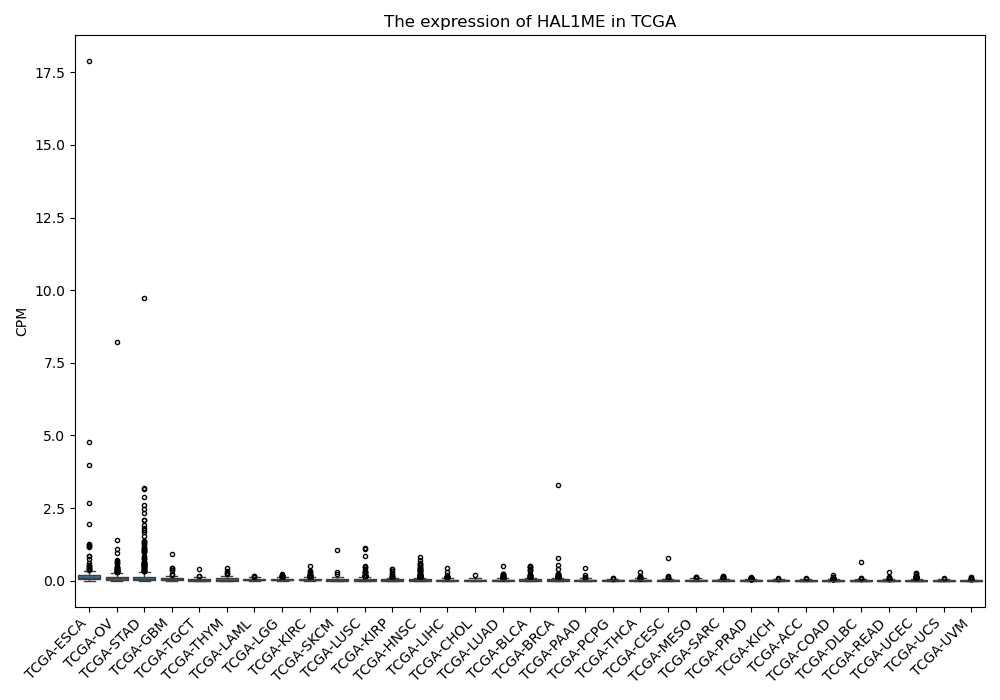
HAL1ME
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000157 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2465 |
| Kimura value | 32.00 |
| Tau index | 0.8302 |
| Description | L1 retrotransposon, HAL1ME subfamily |
| Comment | Full ORF1 between ~724-1707. |
| Sequence |
GGCTTCCGGTTCAAGATGGCGGACTGAGCACACGCGTCTACCTCCCCTCCCTCCCAAAATCCCATTGAAATGACAGAAAAGATATTTTTAAAAGAATGAAACCATAACAGCGCTGGAAAACAAGAAAGGGTGCCATCAGCGGACCAGAAATTTTGAGGAATTCCTGGAAGATAGAAAGCAGATGGGATCGGATTGACGGAGAAANCAGAGCAGAGGAAACCGCAGCCCAAAACACCGGAGGTAAGAGCGGTTCTTCCCAAGGGAGCCCCGGAGAAGCTCCAAGCTCGGAGTCAACAAATACAAAGAGCAGGAGTGGGCCGTGGGGCAATCATCAGGGTAATTAATTAAAGAACTGCATTCGGAACAGCTGGGCCAGTGGGTCCTCCCTCCCCCTCTCCCGCGTANACANAGAGCAGCTGGCAGCAGCGGCTTTTGCCCCCAGGCTAAAGCCCGAGAACTGCTCTCTAAAGAAAGTGGGCGATCTGCCTGGGGAGGGCTCCTAGTGTGGGTGTTGGGGCCTGAGAGAGAAGCAGAGCAGAGCTTGAGGTAACTCCGGACCTCCACAACAGACACGGAGGGGAAAAAGGAAAGGGAAGAGGCAGCTCCGCCCAGGAGTGAAATACACTAAAGCGCTCCACCCCCAGAGTAAAGCCCTCCTTACTTTGGCAATTGTGGGGGTCCCCAGCCTGCCTGCCTGCTCCCTGCCCACGCACCCTAAAGGGAAGCCTGCCTGTCGACACGCCCCGCCCACTTACACAGGAGCCCGCAGTCAGCCCTCCCATTCAAGGGAGAGGCCTGTTGGGGAAACAGACCTACACAACTGCAGAGGAAACCCACGCCAGCTACCTATACTAATCAACCTAGTCCTTCATTCATAAATATGAACGGACAACCAAGGATCACCAGACATTTGAGGAAAACCAACAGCATGAAAGAGAAGGACCAAGATGAACAAACAGAACAACTGACCCCGGAGGAAACAGAGATAATTCAGGGAACAGAAGAGAACTTTAAAAAAATTCTAATTAGTATCCTCAGAGAGATTCGAGAAGATATTGCATCCATAAAACAAGAACAGGNTGCTATGAAAAAGAAGCAATCAGAGAACAAGAAAGAGTTCTTGGAAATTAAAAATATGATTGCCAAAATNAAAAATTCAATAGAAGGGTTGGAAGATAAAGTCGAGGAAATCTCCCAGAACGTAGAGCAAAAAGACAAAGAGATGGAAAATATGAGAGAAAAGTTAAGAGACATGGAGGATAGATCCAGGAGGTCCAACATCCATCTAATAGGAGTTCCAGAAGGAGAGAACAGAGANAATGGAGGGGAGGAAATAATCAAAGAAATAATAGAAGAAAATTTCCCNGAGCTGAAGAAAGACACGAGTCTTCAGATTGAAAGGGCCCACCGAGTGCCGAGCAGGATGAATGAAAAAAGACCCACACCTAGACACATCNTNGTGAAATTTCAGAACNCCAAGGATAAAGAGAAAATCCTAAAAGCTTCCAGAGAGAAAAAACAGGTTACCTACAAAGGAACGAGAATCAGACTGGCATCAGACTTCTCATCAGCAACACTGGATGCTAGAAGACAATGGAGCAATGTCTTCAAAGTTCTGAGGGAAAATNATTTTGAACCTAGAATTCTATACCCAGCCAAACTATCAATCAAGTGTGAGGGCAAAATAAAGACATTTTCAGACATGCAAGGACTCAGAAAGTTTACCNCCCACGTACCCTNTNTGAAAAAATTACTCGAGGATGTACTCCAGCAAAACGAAAAAGGAATCCAAGAAAGAGGANGACATGGGATACAAGAAACAGTGGCAGCTGAGTATGACCCAGAAGAAGTGAAGGAAGCTAAAAAGAAATTAAAAGAGAATCCATTTGACCTTGACGCTTGGAANATTCTCCTTCGAGCGGCANAGGTTTAGTGACATAGGATTACATTTCCTTCTCTATGGGTCCAATCACACTACTTGGTTCTGCAGTGAATAATATTTACATAATCATAATANTGTAAATGCTGTTTATTGGTTTTCAATTTTTAGAATCAACCTATAGACAAAGCACGGAAGACTTAATTATAGTTACAGAACAGAATGTAAATGTTATAAACCTTGACAATGTAAAAATAATANAATAACNAAAATTGGGAGGTAGAAGGGGGAAGAGAGGTGGAAGGAAGTGTAAGNGCGCTAANTTCCTCATCTTNCATAGCGGGGAGTCAANAGATACTGTCTAAAGTTGATAAATCAAGAAATAGAGGTNTAAGTATATTATTTAAAGTTATAAAGGTAACCANTAGAAGAACTAAAAATAANTAACAGTNAAAAGTGGTTGCCTCTGGGGAGTGGGATNGAGATGGAAGAGGGNAGGGANAGGGACTACTTTTCATTTTATACCCTTCTGTACTGTTTGAATTTTTTNNACCATGTGCATGTATTACTTTTATAAAAATAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HAL1ME | RVE7 | 80 | 88 | - | 16.38 | AAAATATCT |
| HAL1ME | TCP23 | 1401 | 1408 | + | 16.36 | GGGCCCAC |
| HAL1ME | ESRRA | 1888 | 1896 | - | 16.33 | TCAAGGTCA |
| HAL1ME | ZFP42 | 12 | 24 | + | 16.31 | CAAGATGGCGGAC |
| HAL1ME | KLF15 | 742 | 749 | + | 16.27 | CCCCGCCC |
| HAL1ME | DOF5.1 | 581 | 599 | + | 16.26 | AAAAAGGAAAGGGAAGAGG |
| HAL1ME | RVE6 | 80 | 88 | + | 16.23 | AGATATTTT |
| HAL1ME | RVE5 | 80 | 88 | + | 16.21 | AGATATTTT |
| HAL1ME | KLF5 | 741 | 750 | + | 16.20 | GCCCCGCCCA |
| HAL1ME | SP8 | 742 | 752 | + | 16.20 | CCCCGCCCACT |
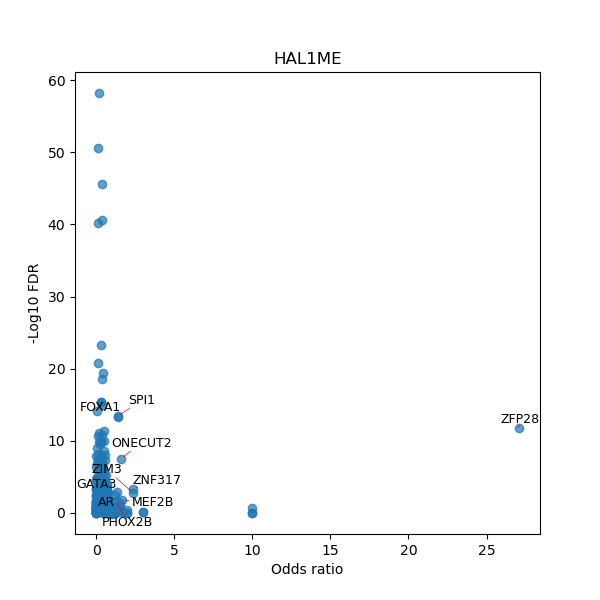
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.