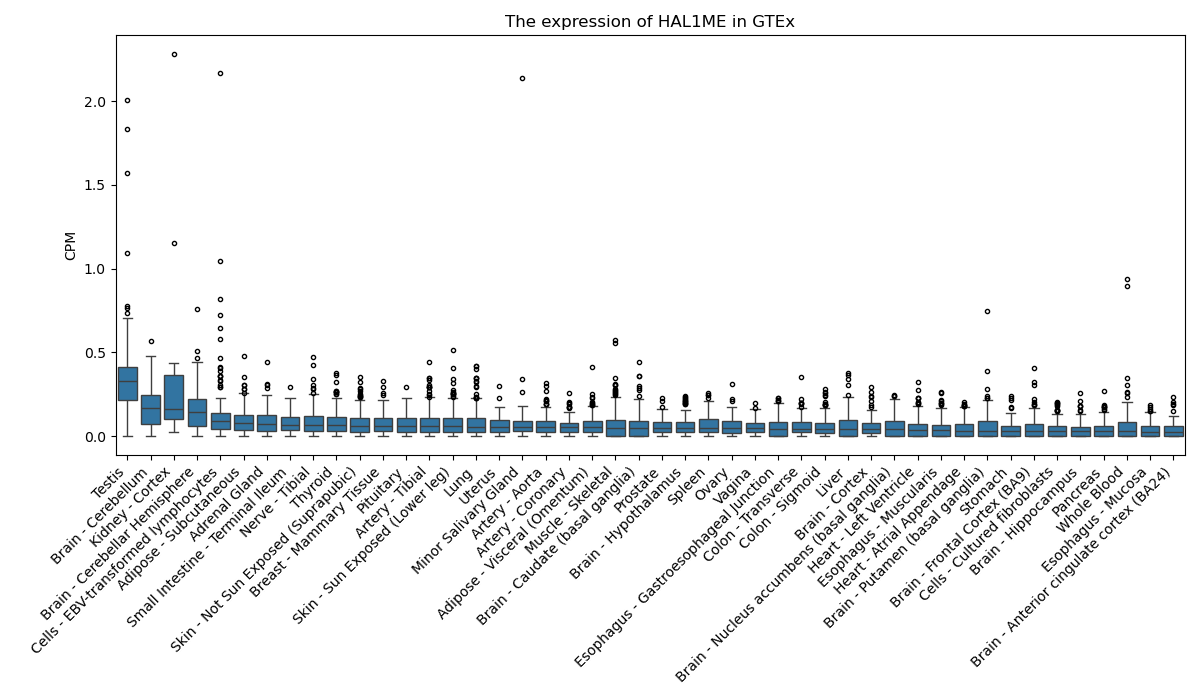
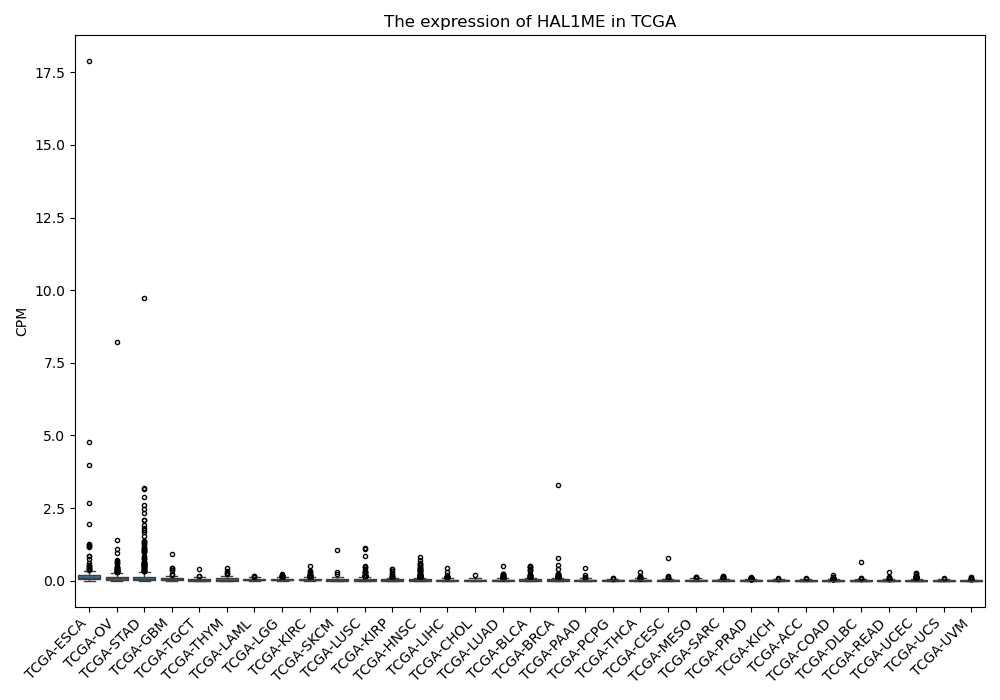
HAL1ME
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000157 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2465 |
| Kimura value | 32.00 |
| Tau index | 0.8302 |
| Description | L1 retrotransposon, HAL1ME subfamily |
| Comment | Full ORF1 between ~724-1707. |
| Sequence |
GGCTTCCGGTTCAAGATGGCGGACTGAGCACACGCGTCTACCTCCCCTCCCTCCCAAAATCCCATTGAAATGACAGAAAAGATATTTTTAAAAGAATGAAACCATAACAGCGCTGGAAAACAAGAAAGGGTGCCATCAGCGGACCAGAAATTTTGAGGAATTCCTGGAAGATAGAAAGCAGATGGGATCGGATTGACGGAGAAANCAGAGCAGAGGAAACCGCAGCCCAAAACACCGGAGGTAAGAGCGGTTCTTCCCAAGGGAGCCCCGGAGAAGCTCCAAGCTCGGAGTCAACAAATACAAAGAGCAGGAGTGGGCCGTGGGGCAATCATCAGGGTAATTAATTAAAGAACTGCATTCGGAACAGCTGGGCCAGTGGGTCCTCCCTCCCCCTCTCCCGCGTANACANAGAGCAGCTGGCAGCAGCGGCTTTTGCCCCCAGGCTAAAGCCCGAGAACTGCTCTCTAAAGAAAGTGGGCGATCTGCCTGGGGAGGGCTCCTAGTGTGGGTGTTGGGGCCTGAGAGAGAAGCAGAGCAGAGCTTGAGGTAACTCCGGACCTCCACAACAGACACGGAGGGGAAAAAGGAAAGGGAAGAGGCAGCTCCGCCCAGGAGTGAAATACACTAAAGCGCTCCACCCCCAGAGTAAAGCCCTCCTTACTTTGGCAATTGTGGGGGTCCCCAGCCTGCCTGCCTGCTCCCTGCCCACGCACCCTAAAGGGAAGCCTGCCTGTCGACACGCCCCGCCCACTTACACAGGAGCCCGCAGTCAGCCCTCCCATTCAAGGGAGAGGCCTGTTGGGGAAACAGACCTACACAACTGCAGAGGAAACCCACGCCAGCTACCTATACTAATCAACCTAGTCCTTCATTCATAAATATGAACGGACAACCAAGGATCACCAGACATTTGAGGAAAACCAACAGCATGAAAGAGAAGGACCAAGATGAACAAACAGAACAACTGACCCCGGAGGAAACAGAGATAATTCAGGGAACAGAAGAGAACTTTAAAAAAATTCTAATTAGTATCCTCAGAGAGATTCGAGAAGATATTGCATCCATAAAACAAGAACAGGNTGCTATGAAAAAGAAGCAATCAGAGAACAAGAAAGAGTTCTTGGAAATTAAAAATATGATTGCCAAAATNAAAAATTCAATAGAAGGGTTGGAAGATAAAGTCGAGGAAATCTCCCAGAACGTAGAGCAAAAAGACAAAGAGATGGAAAATATGAGAGAAAAGTTAAGAGACATGGAGGATAGATCCAGGAGGTCCAACATCCATCTAATAGGAGTTCCAGAAGGAGAGAACAGAGANAATGGAGGGGAGGAAATAATCAAAGAAATAATAGAAGAAAATTTCCCNGAGCTGAAGAAAGACACGAGTCTTCAGATTGAAAGGGCCCACCGAGTGCCGAGCAGGATGAATGAAAAAAGACCCACACCTAGACACATCNTNGTGAAATTTCAGAACNCCAAGGATAAAGAGAAAATCCTAAAAGCTTCCAGAGAGAAAAAACAGGTTACCTACAAAGGAACGAGAATCAGACTGGCATCAGACTTCTCATCAGCAACACTGGATGCTAGAAGACAATGGAGCAATGTCTTCAAAGTTCTGAGGGAAAATNATTTTGAACCTAGAATTCTATACCCAGCCAAACTATCAATCAAGTGTGAGGGCAAAATAAAGACATTTTCAGACATGCAAGGACTCAGAAAGTTTACCNCCCACGTACCCTNTNTGAAAAAATTACTCGAGGATGTACTCCAGCAAAACGAAAAAGGAATCCAAGAAAGAGGANGACATGGGATACAAGAAACAGTGGCAGCTGAGTATGACCCAGAAGAAGTGAAGGAAGCTAAAAAGAAATTAAAAGAGAATCCATTTGACCTTGACGCTTGGAANATTCTCCTTCGAGCGGCANAGGTTTAGTGACATAGGATTACATTTCCTTCTCTATGGGTCCAATCACACTACTTGGTTCTGCAGTGAATAATATTTACATAATCATAATANTGTAAATGCTGTTTATTGGTTTTCAATTTTTAGAATCAACCTATAGACAAAGCACGGAAGACTTAATTATAGTTACAGAACAGAATGTAAATGTTATAAACCTTGACAATGTAAAAATAATANAATAACNAAAATTGGGAGGTAGAAGGGGGAAGAGAGGTGGAAGGAAGTGTAAGNGCGCTAANTTCCTCATCTTNCATAGCGGGGAGTCAANAGATACTGTCTAAAGTTGATAAATCAAGAAATAGAGGTNTAAGTATATTATTTAAAGTTATAAAGGTAACCANTAGAAGAACTAAAAATAANTAACAGTNAAAAGTGGTTGCCTCTGGGGAGTGGGATNGAGATGGAAGAGGGNAGGGANAGGGACTACTTTTCATTTTATACCCTTCTGTACTGTTTGAATTTTTTNNACCATGTGCATGTATTACTTTTATAAAAATAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HAL1ME | ZNF558 | 1368 | 1396 | + | 23.21 | AGCTGAAGAAAGACACGAGTCTTCAGATT |
| HAL1ME | PRDM9 | 2164 | 2183 | + | 19.65 | AGGGGGAAGAGAGGTGGAAG |
| HAL1ME | Spps | 741 | 751 | + | 19.10 | GCCCCGCCCAC |
| HAL1ME | ZNF93 | 416 | 429 | + | 18.71 | GCTGGCAGCAGCGG |
| HAL1ME | phol | 12 | 21 | - | 18.44 | CGCCATCTTG |
| HAL1ME | SP3 | 741 | 751 | + | 18.02 | GCCCCGCCCAC |
| HAL1ME | IDD4 | 1207 | 1221 | + | 18.02 | GCAAAAAGACAAAGA |
| HAL1ME | IDD6 | 1206 | 1221 | - | 17.89 | TCTTTGTCTTTTTGCT |
| HAL1ME | IDD6 | 1206 | 1221 | - | 17.87 | TCTTTGTCTTTTTGCT |
| HAL1ME | ESRRB | 1887 | 1896 | - | 17.53 | TCAAGGTCAA |
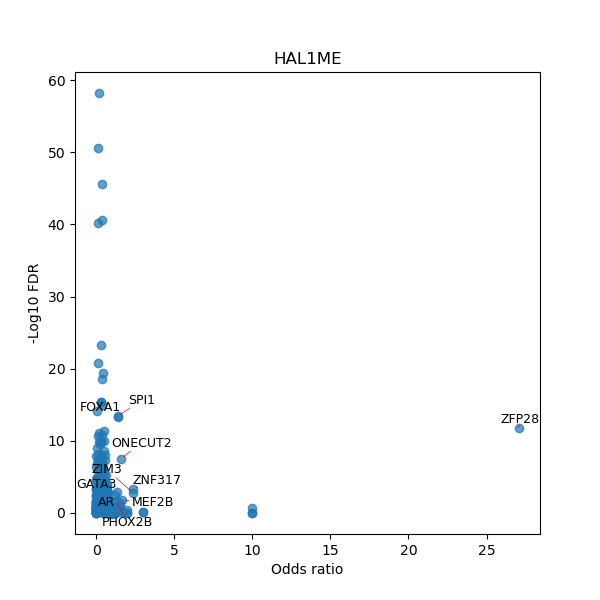
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.