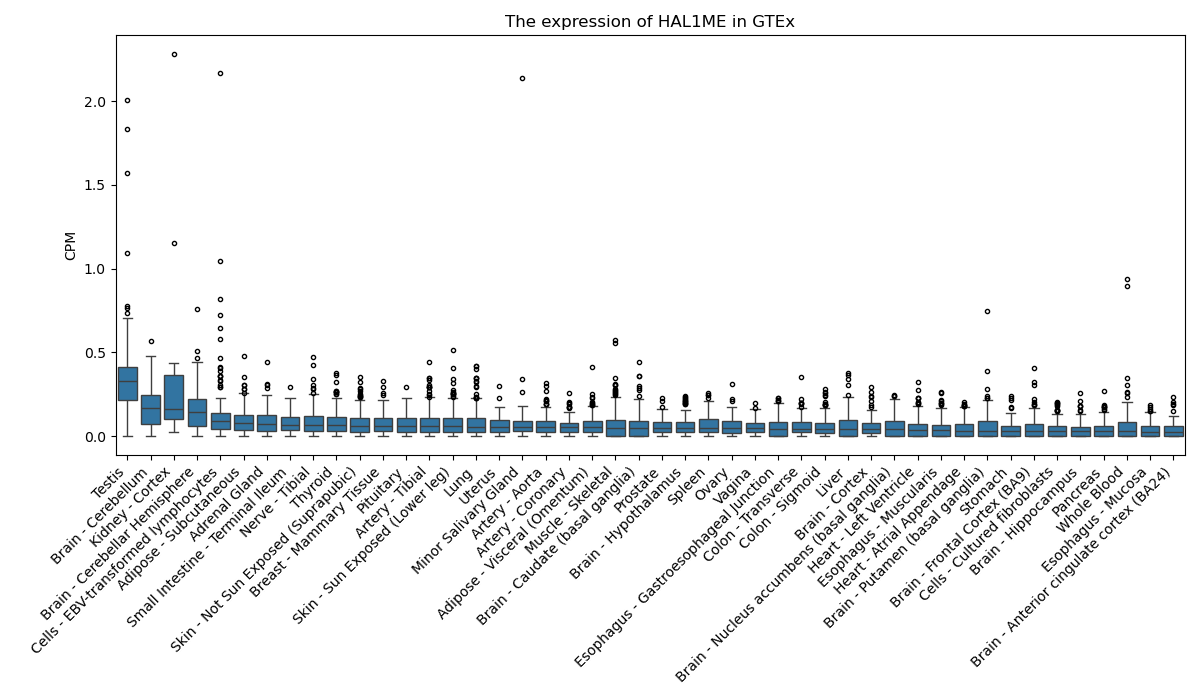
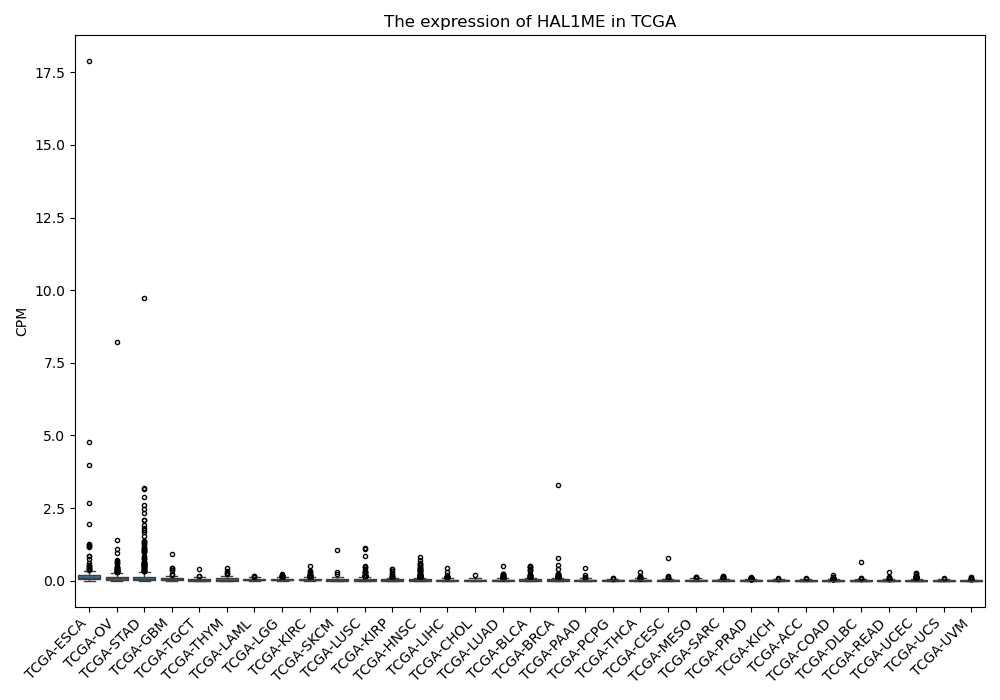
HAL1ME
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000157 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2465 |
| Kimura value | 32.00 |
| Tau index | 0.8302 |
| Description | L1 retrotransposon, HAL1ME subfamily |
| Comment | Full ORF1 between ~724-1707. |
| Sequence |
GGCTTCCGGTTCAAGATGGCGGACTGAGCACACGCGTCTACCTCCCCTCCCTCCCAAAATCCCATTGAAATGACAGAAAAGATATTTTTAAAAGAATGAAACCATAACAGCGCTGGAAAACAAGAAAGGGTGCCATCAGCGGACCAGAAATTTTGAGGAATTCCTGGAAGATAGAAAGCAGATGGGATCGGATTGACGGAGAAANCAGAGCAGAGGAAACCGCAGCCCAAAACACCGGAGGTAAGAGCGGTTCTTCCCAAGGGAGCCCCGGAGAAGCTCCAAGCTCGGAGTCAACAAATACAAAGAGCAGGAGTGGGCCGTGGGGCAATCATCAGGGTAATTAATTAAAGAACTGCATTCGGAACAGCTGGGCCAGTGGGTCCTCCCTCCCCCTCTCCCGCGTANACANAGAGCAGCTGGCAGCAGCGGCTTTTGCCCCCAGGCTAAAGCCCGAGAACTGCTCTCTAAAGAAAGTGGGCGATCTGCCTGGGGAGGGCTCCTAGTGTGGGTGTTGGGGCCTGAGAGAGAAGCAGAGCAGAGCTTGAGGTAACTCCGGACCTCCACAACAGACACGGAGGGGAAAAAGGAAAGGGAAGAGGCAGCTCCGCCCAGGAGTGAAATACACTAAAGCGCTCCACCCCCAGAGTAAAGCCCTCCTTACTTTGGCAATTGTGGGGGTCCCCAGCCTGCCTGCCTGCTCCCTGCCCACGCACCCTAAAGGGAAGCCTGCCTGTCGACACGCCCCGCCCACTTACACAGGAGCCCGCAGTCAGCCCTCCCATTCAAGGGAGAGGCCTGTTGGGGAAACAGACCTACACAACTGCAGAGGAAACCCACGCCAGCTACCTATACTAATCAACCTAGTCCTTCATTCATAAATATGAACGGACAACCAAGGATCACCAGACATTTGAGGAAAACCAACAGCATGAAAGAGAAGGACCAAGATGAACAAACAGAACAACTGACCCCGGAGGAAACAGAGATAATTCAGGGAACAGAAGAGAACTTTAAAAAAATTCTAATTAGTATCCTCAGAGAGATTCGAGAAGATATTGCATCCATAAAACAAGAACAGGNTGCTATGAAAAAGAAGCAATCAGAGAACAAGAAAGAGTTCTTGGAAATTAAAAATATGATTGCCAAAATNAAAAATTCAATAGAAGGGTTGGAAGATAAAGTCGAGGAAATCTCCCAGAACGTAGAGCAAAAAGACAAAGAGATGGAAAATATGAGAGAAAAGTTAAGAGACATGGAGGATAGATCCAGGAGGTCCAACATCCATCTAATAGGAGTTCCAGAAGGAGAGAACAGAGANAATGGAGGGGAGGAAATAATCAAAGAAATAATAGAAGAAAATTTCCCNGAGCTGAAGAAAGACACGAGTCTTCAGATTGAAAGGGCCCACCGAGTGCCGAGCAGGATGAATGAAAAAAGACCCACACCTAGACACATCNTNGTGAAATTTCAGAACNCCAAGGATAAAGAGAAAATCCTAAAAGCTTCCAGAGAGAAAAAACAGGTTACCTACAAAGGAACGAGAATCAGACTGGCATCAGACTTCTCATCAGCAACACTGGATGCTAGAAGACAATGGAGCAATGTCTTCAAAGTTCTGAGGGAAAATNATTTTGAACCTAGAATTCTATACCCAGCCAAACTATCAATCAAGTGTGAGGGCAAAATAAAGACATTTTCAGACATGCAAGGACTCAGAAAGTTTACCNCCCACGTACCCTNTNTGAAAAAATTACTCGAGGATGTACTCCAGCAAAACGAAAAAGGAATCCAAGAAAGAGGANGACATGGGATACAAGAAACAGTGGCAGCTGAGTATGACCCAGAAGAAGTGAAGGAAGCTAAAAAGAAATTAAAAGAGAATCCATTTGACCTTGACGCTTGGAANATTCTCCTTCGAGCGGCANAGGTTTAGTGACATAGGATTACATTTCCTTCTCTATGGGTCCAATCACACTACTTGGTTCTGCAGTGAATAATATTTACATAATCATAATANTGTAAATGCTGTTTATTGGTTTTCAATTTTTAGAATCAACCTATAGACAAAGCACGGAAGACTTAATTATAGTTACAGAACAGAATGTAAATGTTATAAACCTTGACAATGTAAAAATAATANAATAACNAAAATTGGGAGGTAGAAGGGGGAAGAGAGGTGGAAGGAAGTGTAAGNGCGCTAANTTCCTCATCTTNCATAGCGGGGAGTCAANAGATACTGTCTAAAGTTGATAAATCAAGAAATAGAGGTNTAAGTATATTATTTAAAGTTATAAAGGTAACCANTAGAAGAACTAAAAATAANTAACAGTNAAAAGTGGTTGCCTCTGGGGAGTGGGATNGAGATGGAAGAGGGNAGGGANAGGGACTACTTTTCATTTTATACCCTTCTGTACTGTTTGAATTTTTTNNACCATGTGCATGTATTACTTTTATAAAAATAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HAL1ME | BPC5 | 1225 | 1254 | + | -40.71 | GGAAAATATGAGAGAAAAGTTAAGAGACAT |
| HAL1ME | BPC5 | 1227 | 1256 | + | -41.38 | AAAATATGAGAGAAAAGTTAAGAGACATGG |
| HAL1ME | BPC5 | 932 | 961 | + | -42.51 | AAAGAGAAGGACCAAGATGAACAAACAGAA |
| HAL1ME | BPC5 | 1207 | 1236 | + | -43.57 | GCAAAAAGACAAAGAGATGGAAAATATGAG |
| HAL1ME | BPC5 | 566 | 595 | + | -44.54 | ACAGACACGGAGGGGAAAAAGGAAAGGGAA |
| HAL1ME | BPC5 | 2149 | 2178 | + | -47.70 | AAATTGGGAGGTAGAAGGGGGAAGAGAGGT |
| HAL1ME | BPC5 | 1671 | 1700 | + | -47.88 | AGTGTGAGGGCAAAATAAAGACATTTTCAG |
| HAL1ME | BPC5 | 1229 | 1258 | + | -49.20 | AATATGAGAGAAAAGTTAAGAGACATGGAG |
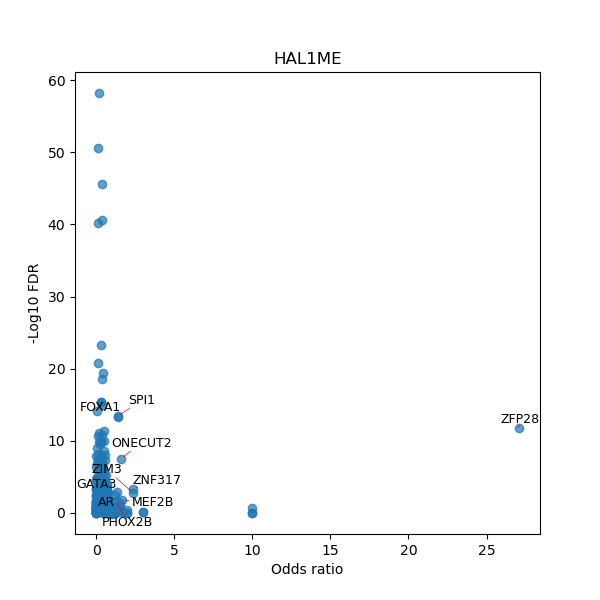
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.