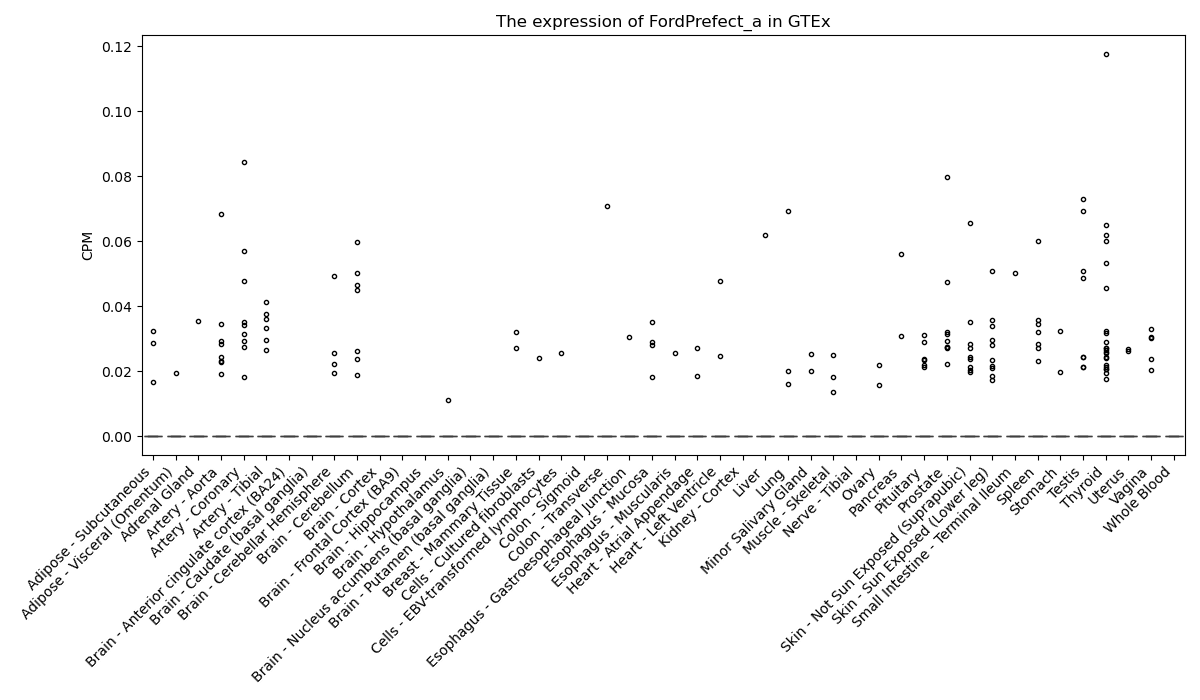
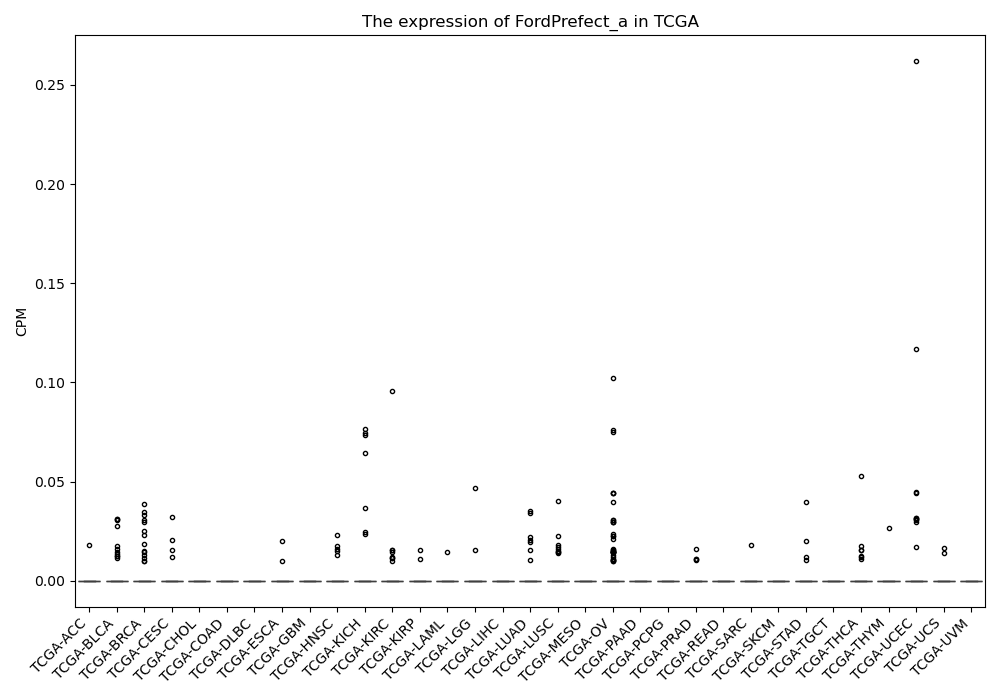
FordPrefect_a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000146 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 508 |
| Kimura value | 25.22 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, FordPrefect_a subfamily (non-autonomous) |
| Comment | FordPrefect_a is an internal deletion product of FordPrefect. It had (at least) 41 CpG dimers, basically representing a mobile CpG island. |
| Sequence |
CAGTACCGCCCTTAGACCCGGGCAAGCGGGGCCCCTGCCCCGGGCCCCGCGCCTCAGGGGACCCCGCGCTTTGGAGTGCCTTCCTCGAAATTTTCTAAGTCCCCCTCCGGTGCCTGGGACCGGCCGGGGCCGGCAGTGCCATCCGGGCGGGGCGCCCCGAGCCCGGACCGGGACCCGCGTGGGCCCCGGTCCCCGTTTCTCCCCTTCGGGGCGCTCTCCGACCCTCTACCCCATGCGTGGGGTCGACCAGATGCCCCGAGGAGCTCCGGGACTCGCGCCTATGGGGTCGTCCCGGGGCCCCGTGGCCGGGCCCCGGTTCCAGGAGGGCGGCCTGGCGAGCGGATCGCTCCCGCTGGCCGGCGCGGTATTTCTTTCGCGGGATCGCGTGAGATTGGGCCGCCAGAATGGTGCTGACACGCTGATTTNGGGTGACTCAAATTGTTTATATAGACAGGGGCCCCGCAAAAAATATTTGCCCGGGGCCCCGCACACCCTAGGGGCGGCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| FordPrefect_a | E2FC | 391 | 405 | + | 2.26 | ATTGGGCCGCCAGAA |
| FordPrefect_a | RXRB | 219 | 232 | - | 1.60 | GGGGTAGAGGGTCG |
| FordPrefect_a | RXRB | 7 | 20 | - | -0.73 | CGGGTCTAAGGGCG |
| FordPrefect_a | PAX9 | 139 | 154 | - | -0.85 | CGCCCCGCCCGGATGG |
| FordPrefect_a | ESR1 | 149 | 163 | + | -1.11 | GGGGCGCCCCGAGCC |
| FordPrefect_a | TCP14 | 179 | 196 | - | -2.80 | ACGGGGACCGGGGCCCAC |
| FordPrefect_a | BCL6B | 369 | 385 | + | -6.89 | TTCTTTCGCGGGATCGC |
| FordPrefect_a | GLIS1 | 59 | 73 | + | -9.42 | GGACCCCGCGCTTTG |
| FordPrefect_a | GLIS1 | 98 | 112 | + | -11.16 | AGTCCCCCTCCGGTG |
| FordPrefect_a | DAL81 | 119 | 137 | + | -52.00 | ACCGGCCGGGGCCGGCAGT |
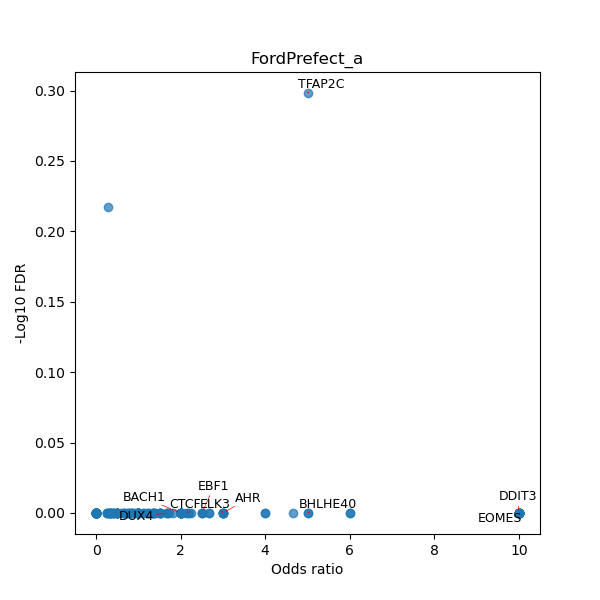
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.