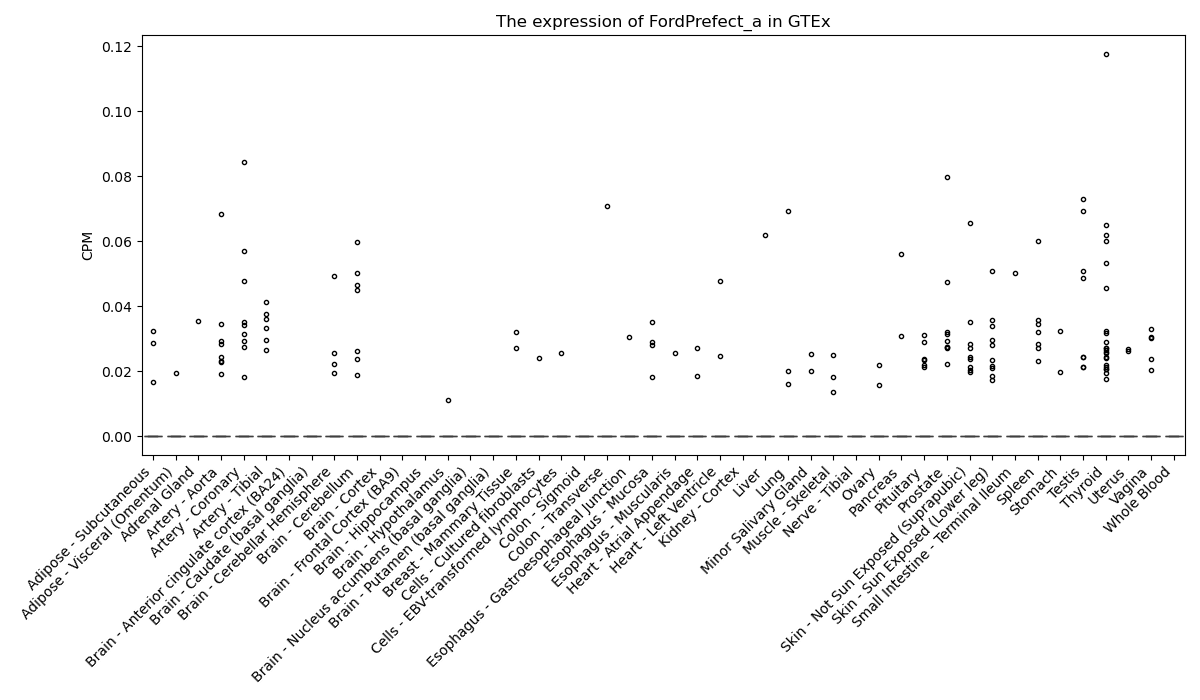
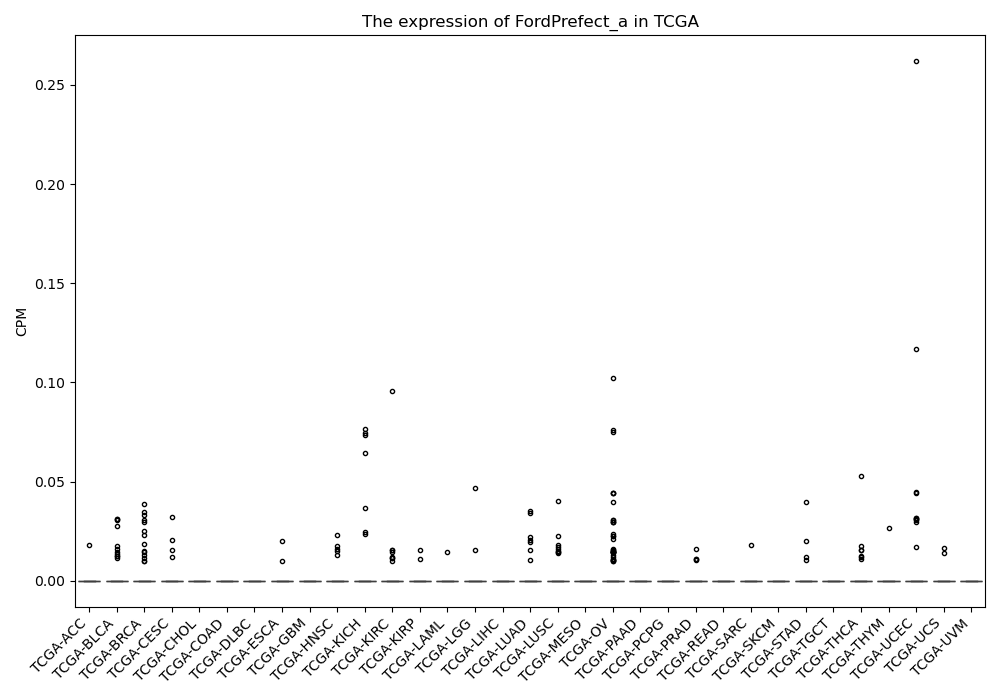
FordPrefect_a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000146 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 508 |
| Kimura value | 25.22 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, FordPrefect_a subfamily (non-autonomous) |
| Comment | FordPrefect_a is an internal deletion product of FordPrefect. It had (at least) 41 CpG dimers, basically representing a mobile CpG island. |
| Sequence |
CAGTACCGCCCTTAGACCCGGGCAAGCGGGGCCCCTGCCCCGGGCCCCGCGCCTCAGGGGACCCCGCGCTTTGGAGTGCCTTCCTCGAAATTTTCTAAGTCCCCCTCCGGTGCCTGGGACCGGCCGGGGCCGGCAGTGCCATCCGGGCGGGGCGCCCCGAGCCCGGACCGGGACCCGCGTGGGCCCCGGTCCCCGTTTCTCCCCTTCGGGGCGCTCTCCGACCCTCTACCCCATGCGTGGGGTCGACCAGATGCCCCGAGGAGCTCCGGGACTCGCGCCTATGGGGTCGTCCCGGGGCCCCGTGGCCGGGCCCCGGTTCCAGGAGGGCGGCCTGGCGAGCGGATCGCTCCCGCTGGCCGGCGCGGTATTTCTTTCGCGGGATCGCGTGAGATTGGGCCGCCAGAATGGTGCTGACACGCTGATTTNGGGTGACTCAAATTGTTTATATAGACAGGGGCCCCGCAAAAAATATTTGCCCGGGGCCCCGCACACCCTAGGGGCGGCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| FordPrefect_a | BAD1 | 477 | 488 | + | 19.90 | CCGGGGCCCCGC |
| FordPrefect_a | BAD1 | 305 | 316 | - | 19.20 | CCGGGGCCCGGC |
| FordPrefect_a | BAD1 | 39 | 50 | + | 18.56 | CCCGGGCCCCGC |
| FordPrefect_a | BAD1 | 306 | 317 | + | 17.20 | CCGGGCCCCGGT |
| FordPrefect_a | BAD1 | 292 | 303 | + | 16.72 | CCGGGGCCCCGT |
| FordPrefect_a | PATZ1 | 143 | 153 | + | 16.65 | CCGGGCGGGGC |
| FordPrefect_a | FOXE1 | 439 | 450 | - | 16.52 | CTATATAAACAA |
| FordPrefect_a | TCP23 | 179 | 186 | - | 16.36 | GGGCCCAC |
| FordPrefect_a | KLF15 | 145 | 152 | - | 16.27 | CCCCGCCC |
| FordPrefect_a | cassava45561.m1 | 179 | 187 | - | 16.09 | GGGGCCCAC |
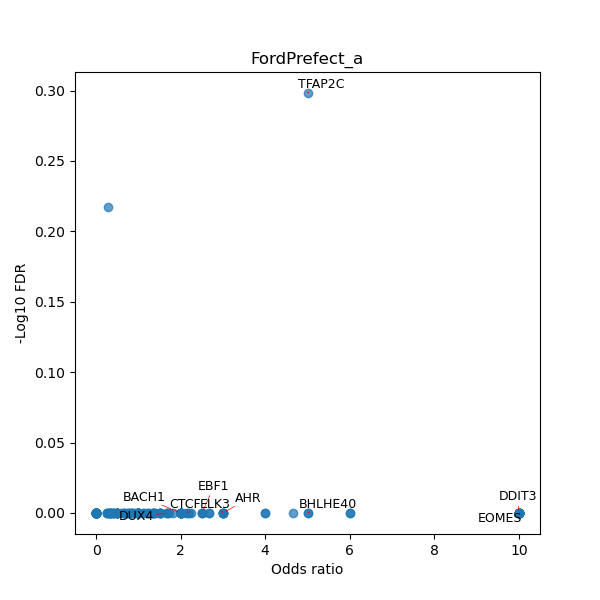
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.