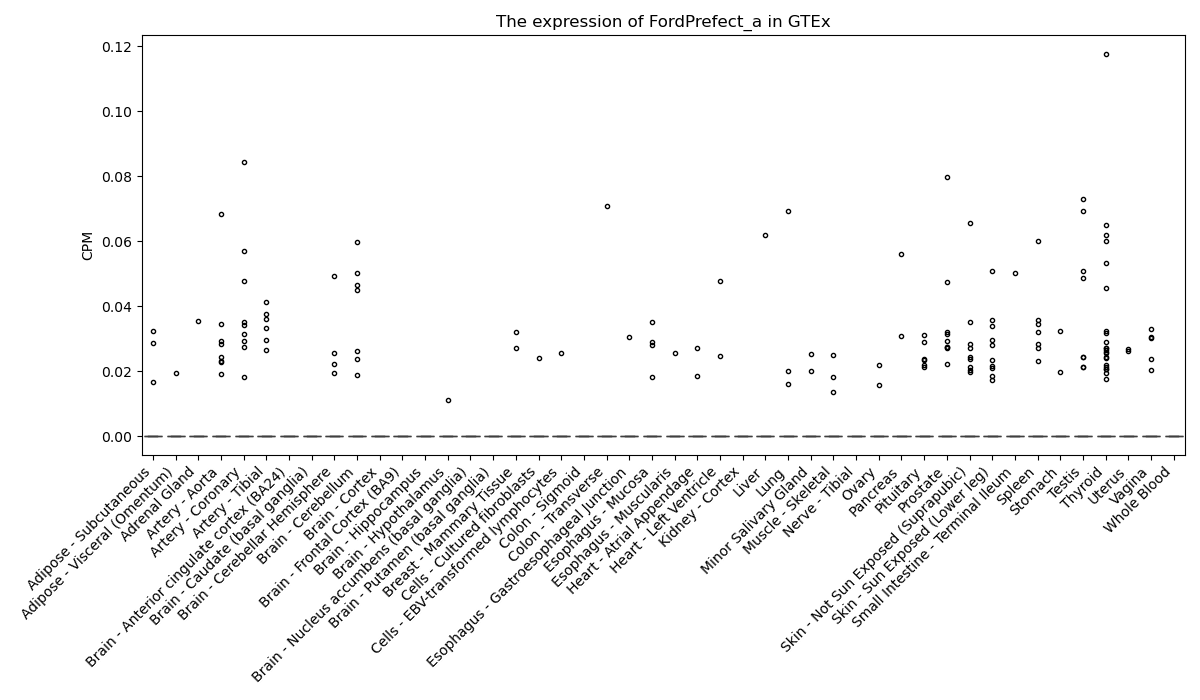
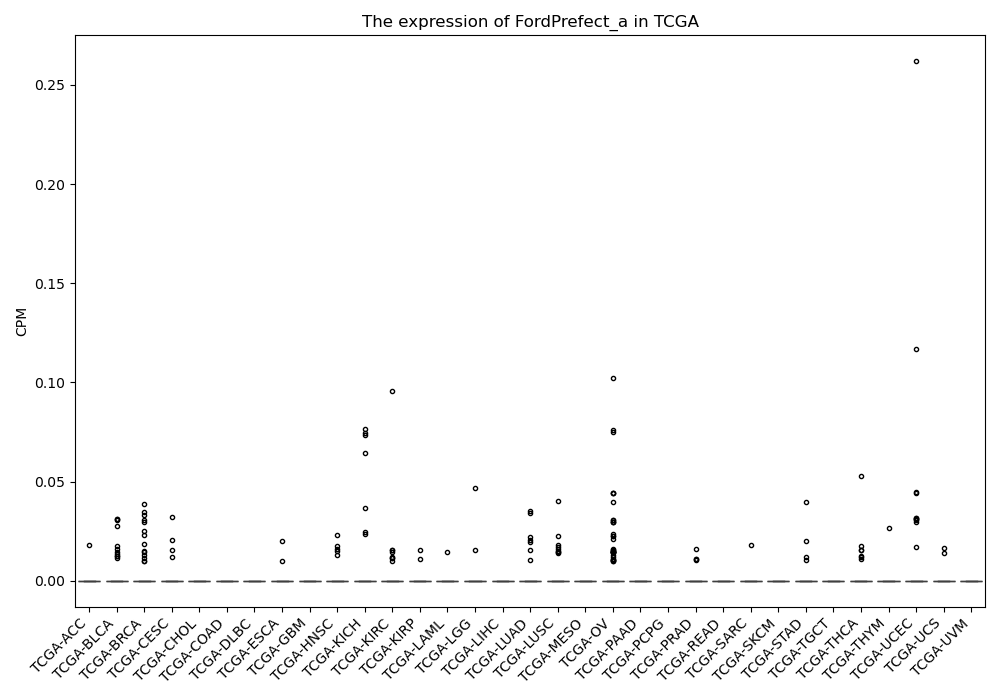
FordPrefect_a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000146 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 508 |
| Kimura value | 25.22 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, FordPrefect_a subfamily (non-autonomous) |
| Comment | FordPrefect_a is an internal deletion product of FordPrefect. It had (at least) 41 CpG dimers, basically representing a mobile CpG island. |
| Sequence |
CAGTACCGCCCTTAGACCCGGGCAAGCGGGGCCCCTGCCCCGGGCCCCGCGCCTCAGGGGACCCCGCGCTTTGGAGTGCCTTCCTCGAAATTTTCTAAGTCCCCCTCCGGTGCCTGGGACCGGCCGGGGCCGGCAGTGCCATCCGGGCGGGGCGCCCCGAGCCCGGACCGGGACCCGCGTGGGCCCCGGTCCCCGTTTCTCCCCTTCGGGGCGCTCTCCGACCCTCTACCCCATGCGTGGGGTCGACCAGATGCCCCGAGGAGCTCCGGGACTCGCGCCTATGGGGTCGTCCCGGGGCCCCGTGGCCGGGCCCCGGTTCCAGGAGGGCGGCCTGGCGAGCGGATCGCTCCCGCTGGCCGGCGCGGTATTTCTTTCGCGGGATCGCGTGAGATTGGGCCGCCAGAATGGTGCTGACACGCTGATTTNGGGTGACTCAAATTGTTTATATAGACAGGGGCCCCGCAAAAAATATTTGCCCGGGGCCCCGCACACCCTAGGGGCGGCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| FordPrefect_a | LjSGA_053525.1 | 179 | 186 | - | 16.05 | GGGCCCAC |
| FordPrefect_a | BAD1 | 40 | 51 | + | 16.01 | CCGGGCCCCGCG |
| FordPrefect_a | PK19717.1 | 179 | 187 | - | 15.77 | GGGGCCCAC |
| FordPrefect_a | KLF7 | 145 | 152 | + | 15.72 | GGGCGGGG |
| FordPrefect_a | OsI_08196 | 179 | 186 | - | 15.69 | GGGCCCAC |
| FordPrefect_a | ARALYDRAFT_493022 | 179 | 186 | - | 15.63 | GGGCCCAC |
| FordPrefect_a | KLF1 | 145 | 152 | + | 15.53 | GGGCGGGG |
| FordPrefect_a | TCP21 | 179 | 189 | + | 15.44 | GTGGGCCCCGG |
| FordPrefect_a | ARALYDRAFT_495258 | 179 | 186 | - | 15.43 | GGGCCCAC |
| FordPrefect_a | ARALYDRAFT_484486 | 179 | 186 | - | 15.43 | GGGCCCAC |
TFBS enrichment in GRCh38
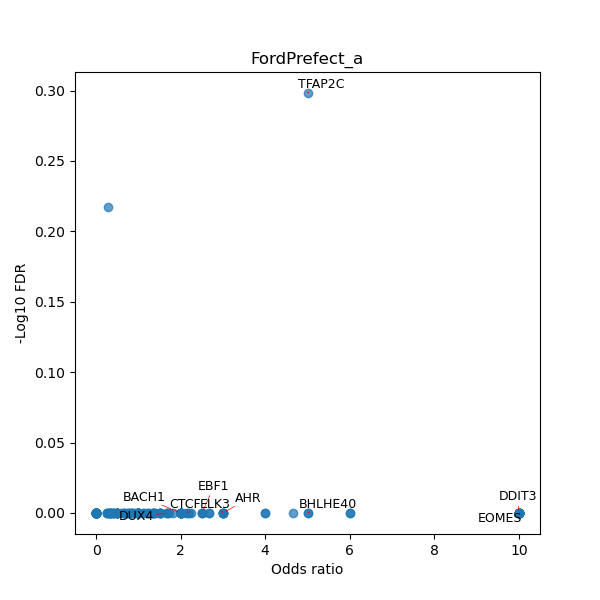
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.