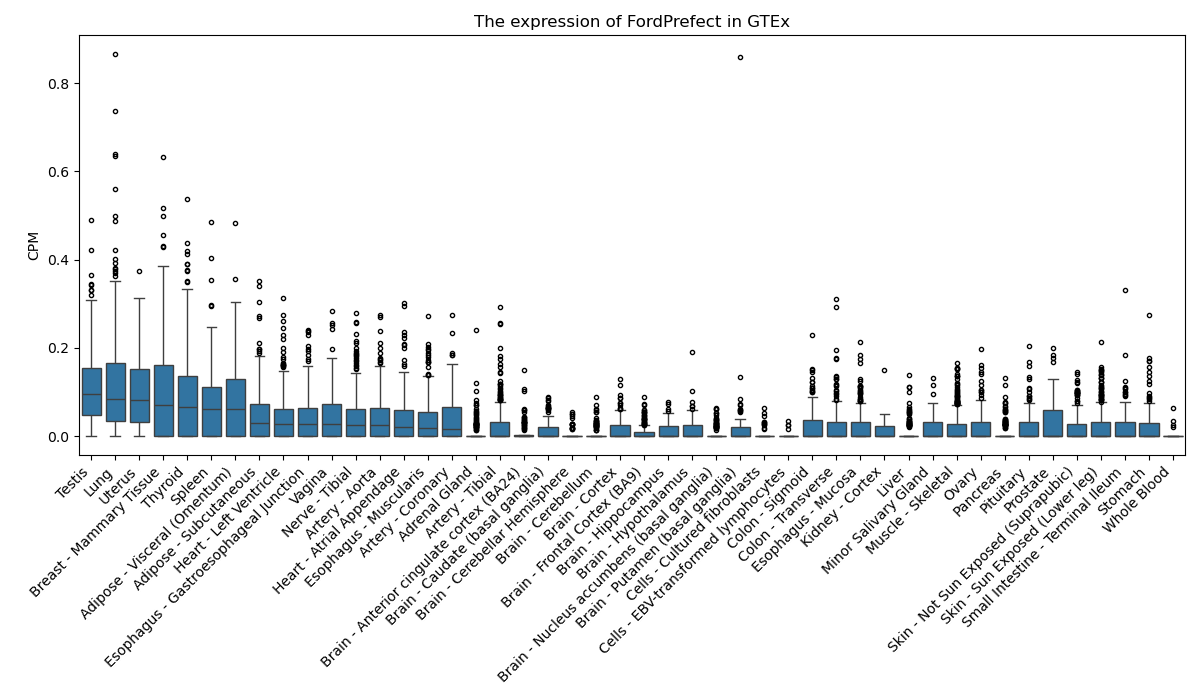
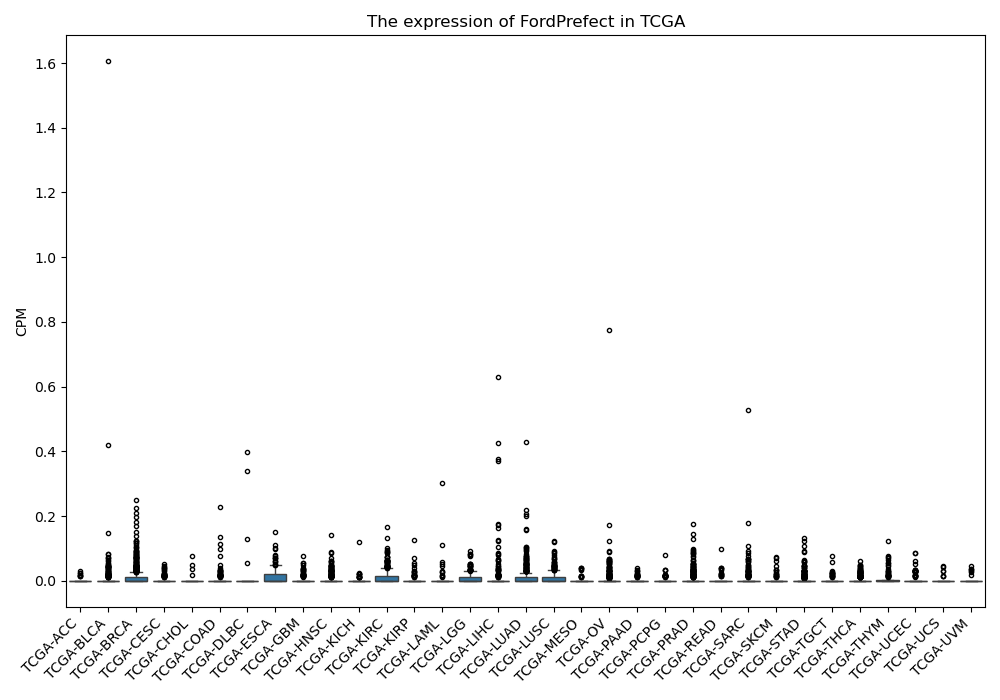
FordPrefect
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000145 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 1683 |
| Kimura value | 23.98 |
| Tau index | 0.8502 |
| Description | hAT-Tip100 DNA transposon, FordPrefect subfamily (non-autonomous) |
| Comment | Has 8bp TSDs. The first 66 bp of FordPrefect are 90% ID, and the last 36 bp are 80% ID to those of Zaphod. FordPrefect does not seem to have had coding capacity and perhaps hitchhiked with Zaphod. |
| Sequence |
CAGTACCGCCCTTAGACCCGGGCAAGCGGGGCCCCTGCCCCGGGCCCCGCGCCTCAGGGGACCCCGTGCTTTGGAGTGCCTTCCTCGAAATTTTCTAAGTCCCCCTCCGGTGCCTGGGACCGGCCGGGGCCGGCAGTGCCATCCGGGCGGGGCGCCCCGAGCCCGGACCGGGACCCGCGTGGGCCCTGGTCCCCGTTTCTCCCCTTCGGGGCGCTCTCCGACCCTCTACCCCGCGCGTGGGGTCGACCAGATGCCCCGAGGAGCTCCGGGACTCGCGCCTATGGGGTCGTCCCGGGGCCCCGTGGCCGGGCCCCGGTTCCAGGAGGGCGGCCTGGCGAGCGGATCGCTCCCGCTGGCCGGCGCGGTATTTCTTTCGCGGGATCGCGCGAGATTGGGCCGCCAGAATGGTGCTGACACGCTGATTTGGGGTGACTCTCACTCACGTCGGACACAGGACGAGTTCAGGGCTCTGGGCTACCGACGGTCCACCGCCGACCCTTGGGCTTGAGCCGCATGTGTGGGCCCATGCGTCGGCTCTCGCCCGTCCGTGTTCCGACCGCGGTGCCGCCTCCGGTCTACAGCACCCGAGGGCGGCGGGGGTGGCGGCAGGCATCCTTTACCCTGTGCGCCTCCCACCGCTGGCACCCAGGGCGGTCACCCCACCCCCTCCGCAGGCTCCGCGCCACGTGTCAGGCAGTCCTCCGGGGGTGGCCGCGCCTATCTCCTCCGAGGGCTTTCGAGACCGTTGCTCCGCAACGCCAACGGGCCCTTCCGATCGATGTCCTCTCTTGCCTCCGATCGATGTGGTGACGTCGTGCTCTCCCGGGTCGGTCTTAAGCCGTGCCGGACGAGGGACGGACATTCCTTGCGCGAATGGGACCGCTCTTCTCGCTCCGCCCACGGGCCCCTCGCCTATCCTCCCCGCTGTGGCGGTGTGTGGAAGGCAGGGGTGCGGTCAACATTGAAAGAGATCACATTCTAGGAATGCAGTGATTACGGCCTAAAGAGTTCAAGAGAAGACATGGTTGGAAGATGTGTTGTTCTACGTTTATGCTATAAAATTCCGAACGGTAAATTTAACATGACCAGAAAACGAATTATCGTTCACATTTTCCTGCATACTCTGGGTAAGACTTGCATTTGTGGTCATCATCAACGAAGCACAGTAACAACCTTTGAGAGAGTCATTGGAAGCCAGTATTCACGGGCGGCACGATGGATGATGCAGCGTCATGAGTAATGATGTAACCAGCATTAAATAAATGGTATTAGGGAACTGCAGAGGCAAGAAGATCTATATTGTTTCAATACAAACAGGTTCCGAAGAGCCATGGCATTGTGAGTAATAACAGCGTTGCTACCTTTTTCTCGCGGTGGGAGATATGAAATTAGCCAGGAACGGCGCATTTGACAATAAAGAACACGAAGAGATGGTTCCTGGACCTGAACAGGAAGAGATGGTGCCTGGACACTACGAAGAATCTTCACGTGCACTGATTGGACAATAAACAAATACGTAAGTACCTCTTCTCTACCCATTATTCTAAATCTTCATCGATAAATCACTATACCTCACATGGGCCCATGAATTTTGTAATACATTTTTAATCAAATTGTTTATATAGACAGGGGCCCCGCAAAAAATATTTGCCCGGGGCCCCGCACACCCTAGGGGCGGCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| FordPrefect | KLF15 | 145 | 152 | - | 16.27 | CCCCGCCC |
| FordPrefect | LjSGA_053525.1 | 179 | 186 | - | 16.05 | GGGCCCAC |
| FordPrefect | LjSGA_053525.1 | 518 | 525 | - | 16.05 | GGGCCCAC |
| FordPrefect | SREBF1 | 654 | 663 | + | 16.05 | GTCACCCCAC |
| FordPrefect | ZmbZIP96 | 681 | 691 | - | 16.01 | GACACGTGGCG |
| FordPrefect | BAD1 | 40 | 51 | + | 16.01 | CCGGGCCCCGCG |
| FordPrefect | Creb3l2 | 682 | 690 | + | 15.96 | GCCACGTGT |
| FordPrefect | ERF057 | 590 | 603 | - | 15.84 | CCACCCCCGCCGCC |
| FordPrefect | KLF7 | 145 | 152 | + | 15.72 | GGGCGGGG |
| FordPrefect | ZBED4 | 888 | 897 | + | 15.72 | CTCGCTCCGC |
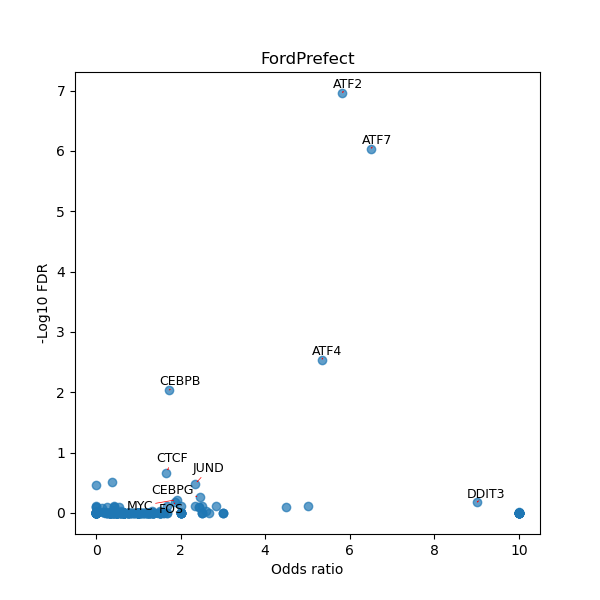
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.