Eutr2B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001264 |
|---|---|
| TE superfamily | MuDR |
| TE class | DNA |
| Species | Eutheria |
| Length | 1699 |
| Kimura value | 25.24 |
| Tau index | 0.0000 |
| Description | Eutr2B subfamily |
| Comment | Pos 13-125 bp of the 250-300 bp long TIRs are ~8 copies of ATTCGCCGAAARYCA. Full TIRs are shared with the shorter element named Eutr2. The remaining 1200 bp are unrelated. |
| Sequence |
GGCTCTAGGCCAATTCGCCGAAAATCAATTCGCCGAAAGCCAATTCGCCGAAAGATCAATTCGCCGAATGACCAATTCGCCGAAAGCCAATTCGCCGAAAATCAATTTGCCGAATGACCAATTCGCCGAATTTACCAAATTTACCGATTTACCAAAAACTTGTTTCTTTTAACTATTTATAAAGTTTACAGCAATTCGTATTGGATGGGATGGTTTTAACAGCTTTTGAAGATTTCTGCGAAGTTTTAGTTGACCGGTTTTCATTTTGTCTTCATAGCAGCATTTTAAGGAATGTTCAATTTGTCGAAAGCTAAGGGTCATAGGGTGGTTTAGTCTATTTNTGTTAAATATATTTAATATTCAGAAAAGTATATTTTATCATCAGTAACCAAAATTCAGCATTTCATATGGGAATTTTTCAGTTTATACTGATTTCTTTCNAATNTTTTTGAAACTTTTTATNATCCATTTTTATGCTTTTTTNAATTATTTTNAGGATTCTTTCGCCAATTTCTCAAGTAGCATATCAACCTTAGTTTGTTATTTCTAGNAATTAGTAGCCAAAATTTAATATGTGATGTGGTTGCTCCTGTCGCTTCCTAGTTCATTTTACTTACCTATCTTTTAGCATTTTTAGGATTTTTAANANTTAATTTTTGATCCTTCCTTCACAATTTCGTATTTATGGTTAATGTATATCAAATTTGAGGTACTAATCATACGGTCCATTATAGTAATCCCTTCTGGGCATCCTCACTTCCTTTCCGCTGCTTTGAACGGTTTTACCCTTTCATGTCGAATCTTTCTGCAAACTGCTACCAAGTCGACATATCCCAATTGCTCCTTGTTAGAAACCAAAATTTCAGCATTTCATATGTGACGGGATTGTTTCGGCTACTTCTGATTTCTTTCGGATGCTTTTGAATGTCATTTTCTATTTTTTCAAACTTTAANCATTTTTAGTATTCTTTCGCCAATTCCTCAAATGGCATATCAGCCTTAATTTTGTATTTCTAGCAATTAAAATTTTAAATTTCAGCATTTCACATGTGATGGGATTGTTTCGGCCGCTTTTGAGTTCTTTCGCATCCTTTTGAATGTCATTTTTNATTTTTCCAATCTTTAGGCATTTTTACCATTCTTTCACCAATGTCTCAAATGGCATATCAGCCTTAATTTTGTATTTCTAGCAATTAGTAACCAAAATTTCGGCATTTCATATTGATGTGGTTGCTCTGGATCCTTCCTCAGCAATTTCATATTTATGGTGAATGTATATCAGATTTGGGGTACTAATCTTACGCTGAAATTTTGGTTACTGATTATGAAATATACTTTTCTGAATATTAAATATATTCATAAGAAGACTAAACCACCCTATGACCCTTAGCTTTTGACAAATTGAACATTCCTTAAAATGCTGCTATGAAGACAAAATGAAAACCGGTCAACTAAAACTTCGCAGAAATCTTCAAAAGCTGTTAAAACCATCCCATCCAATACGAATTGCTGTAAACTTTATAAATAGTTAAAAGAAACAAGTTTTTGGTAAATCGGTAAATTTGGTAAATTCGGCGAATTGGTCATTCGGCGAATTGATTTTCGGCGAATTGGCTTTCGGCGAATTGGTCATTCGGCGAATTGATTTTCGGCGAATTGGCTTTCGGCGAATTGATTTTCGGCGAATTGGCCCGGAGCC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eutr2B | SOL1 | 1020 | 1034 | + | 18.48 | ATTAAAATTTTAAAT |
| Eutr2B | TSO1 | 1019 | 1033 | + | 17.25 | AATTAAAATTTTAAA |
| Eutr2B | TCX6 | 1020 | 1034 | - | 17.13 | ATTTAAAATTTTAAT |
| Eutr2B | E2FC | 552 | 566 | + | 16.84 | AATTAGTAGCCAAAA |
| Eutr2B | TSO1 | 1021 | 1035 | - | 16.00 | AATTTAAAATTTTAA |
| Eutr2B | SPIB | 754 | 766 | + | 15.84 | TCACTTCCTTTCC |
| Eutr2B | ZKSCAN5 | 753 | 761 | - | 15.83 | GGAAGTGAG |
| Eutr2B | TCX2 | 1020 | 1034 | + | 15.63 | ATTAAAATTTTAAAT |
| Eutr2B | Hmx1 | 1016 | 1024 | + | 15.52 | AGCAATTAA |
| Eutr2B | Hmx3 | 1016 | 1024 | + | 15.27 | AGCAATTAA |
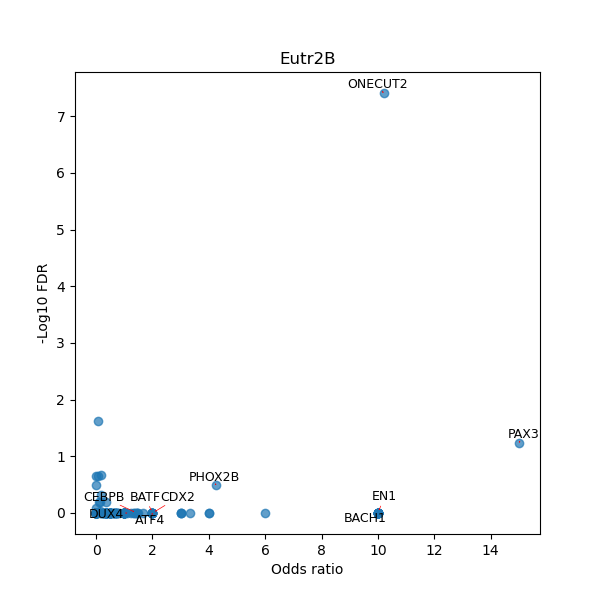
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.