EuthAT-N1a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001194 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 258 |
| Kimura value | 28.43 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, EuthAT-N1a subfamily |
| Comment | - |
| Sequence |
CAGAGCTGTAACTAGGGCGGGGCGACCGGGGCTTTGCCCCAGGGCACCGAAATTTAGAGGGCGCAAANATTTTAAATAATGATTTTAAAAGCTTAGCACCTAGTTAGCAAGAATAATTAGTTAAAATAGGAAGCTACCAGATGGCGCGCATTGTTAATAACACCCATCTGTGAGCCACATCATGAATTACAAAGTTGATTTGAGGGCGCGTTTCGTGCTGCTTGCCCCGGGCGCCAAAATTGCTAGTTACGGCTCTGC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| EuthAT-N1a | KLF7 | 15 | 22 | + | 15.72 | GGGCGGGG |
| EuthAT-N1a | KLF1 | 15 | 22 | + | 15.53 | GGGCGGGG |
| EuthAT-N1a | TFAP2A | 36 | 46 | - | 15.44 | TGCCCTGGGGC |
| EuthAT-N1a | SP4 | 14 | 22 | + | 15.12 | AGGGCGGGG |
| EuthAT-N1a | SP2 | 14 | 22 | + | 14.96 | AGGGCGGGG |
| EuthAT-N1a | KLF12 | 14 | 22 | + | 14.93 | AGGGCGGGG |
| EuthAT-N1a | TBXT | 158 | 173 | + | 14.79 | TAACACCCATCTGTGA |
| EuthAT-N1a | Neurod2 | 164 | 171 | - | 14.69 | ACAGATGG |
| EuthAT-N1a | CTCF | 55 | 64 | + | 14.60 | TAGAGGGCGC |
| EuthAT-N1a | NEUROD1 | 164 | 171 | - | 14.58 | ACAGATGG |
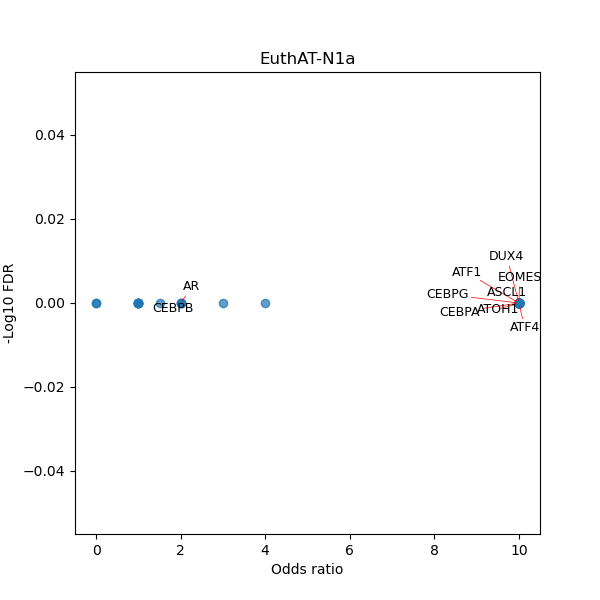
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.