EuthAT-N1a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001194 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 258 |
| Kimura value | 28.43 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, EuthAT-N1a subfamily |
| Comment | - |
| Sequence |
CAGAGCTGTAACTAGGGCGGGGCGACCGGGGCTTTGCCCCAGGGCACCGAAATTTAGAGGGCGCAAANATTTTAAATAATGATTTTAAAAGCTTAGCACCTAGTTAGCAAGAATAATTAGTTAAAATAGGAAGCTACCAGATGGCGCGCATTGTTAATAACACCCATCTGTGAGCCACATCATGAATTACAAAGTTGATTTGAGGGCGCGTTTCGTGCTGCTTGCCCCGGGCGCCAAAATTGCTAGTTACGGCTCTGC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| EuthAT-N1a | CTCF | 138 | 147 | + | 16.72 | CAGATGGCGC |
| EuthAT-N1a | TFAP2B | 36 | 46 | + | 16.67 | GCCCCAGGGCA |
| EuthAT-N1a | TFAP2C | 36 | 46 | + | 16.61 | GCCCCAGGGCA |
| EuthAT-N1a | ZNF682 | 28 | 38 | - | 16.54 | GGCAAAGCCCC |
| EuthAT-N1a | KLF5 | 14 | 23 | - | 16.29 | GCCCCGCCCT |
| EuthAT-N1a | KLF15 | 15 | 22 | - | 16.27 | CCCCGCCC |
| EuthAT-N1a | TFAP2A | 35 | 45 | + | 16.09 | TGCCCCAGGGC |
| EuthAT-N1a | TFAP2B | 35 | 45 | - | 16.00 | GCCCTGGGGCA |
| EuthAT-N1a | Zfp961 | 229 | 236 | + | 15.99 | GGGCGCCA |
| EuthAT-N1a | TFAP2C | 35 | 45 | - | 15.93 | GCCCTGGGGCA |
TFBS enrichment in GRCh38
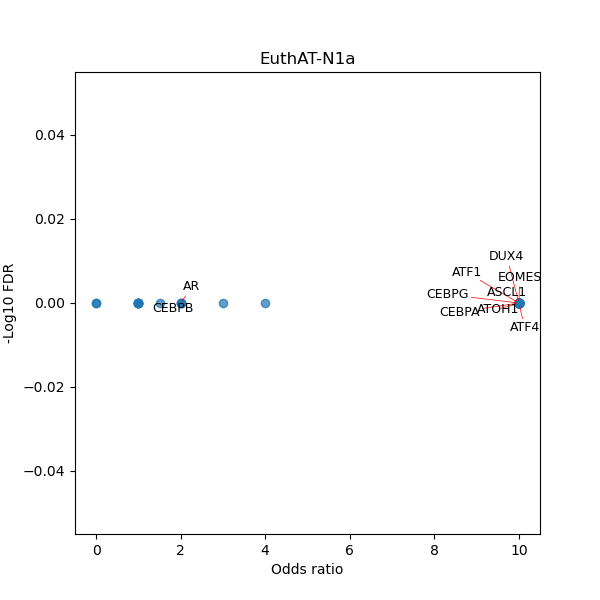
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.