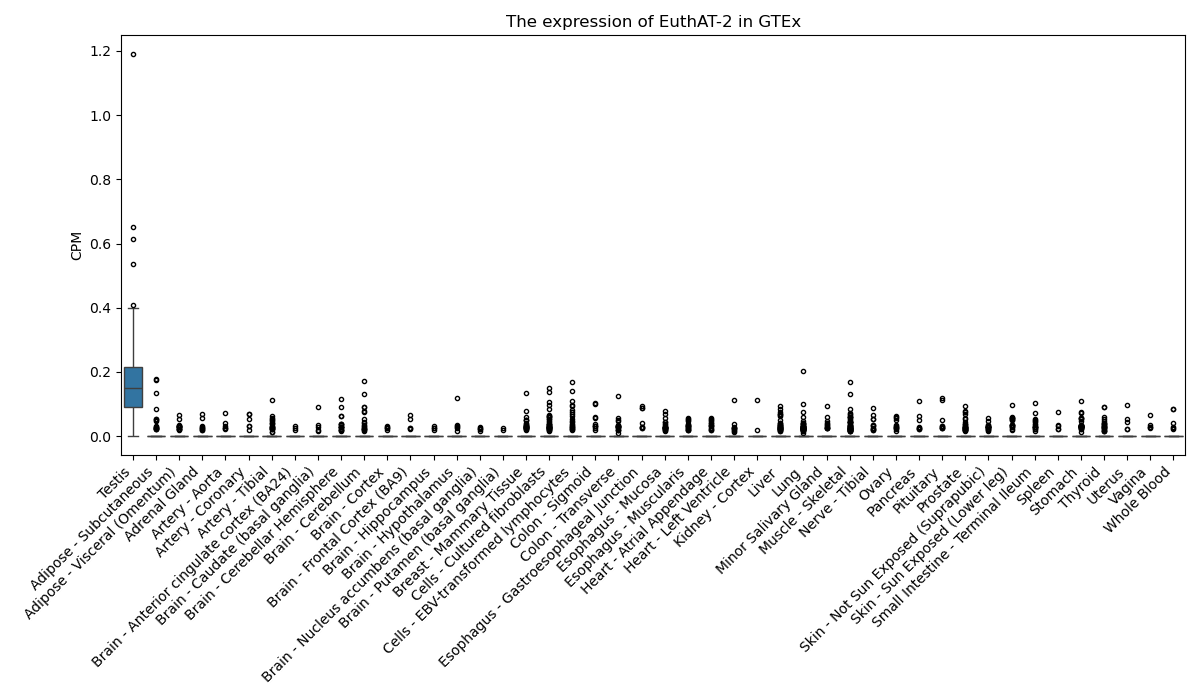
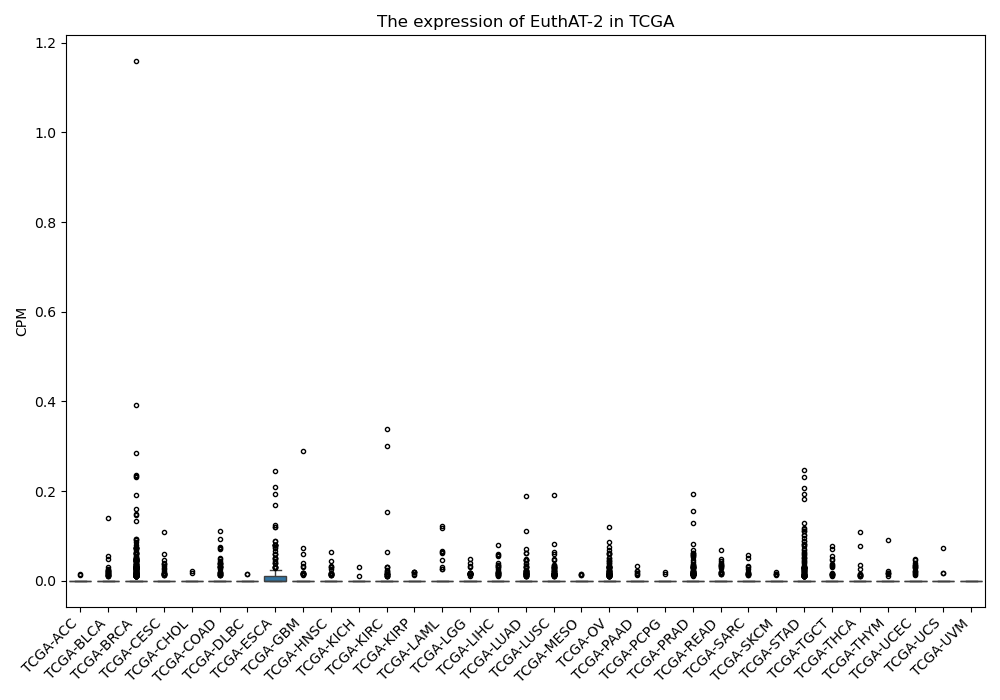
EuthAT-2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001172 |
|---|---|
| TE superfamily | Activator |
| TE class | DNA |
| Species | Eutheria |
| Length | 2747 |
| Kimura value | 25.50 |
| Tau index | 1.0000 |
| Description | Ancient hAT DNA transposon from eutherian mammals. |
| Comment | >74% identical to consensus. It encodes remnants of transposase. No copies in opossum. ORF 727-2562 encoded a transposase closest related to HAT-12_XT_tp of Xenopus and HAT5_CI_tp and HAT1_CI_tp of Ciona. Present at orthologous sites throughout Eutheria, but not in marsupials. |
| Sequence |
TAGGGATGCACCGAAGTTCGGCTTCGGCCTCGGTATTCGGCCAAAATCTGCTATTCGGCTTCGGCTTCGGCCTCGGCCTTGGAAGGCCGAATAGCCGAATGCCTACTTCAACGTCATTTCTGGCATCCCTGAACTTGTGCGTGATCAATGATCGTGTGATCGCGTAGGTTACCATCATATCTAATCAGCGTAACAACGTCAGTAATACGAACAAGGCATGTCATTCGGATTTATTGTTAATATCTGAATATTCACTTTCAAATATAGGTAAGCATTCTAGATGTAACTTGGAGTGAAAAATCTCCTTATGTGCTTTATATGTTGAAATTCTATCAGTCAGTTGAAGTATAAATGTATATTTATTATTGCAAGAGTAAAATGCTGCAATAATTAATGATGATTGTGAAAGCTGTTAATTTTATGTACAAAAACAATTATTAAGAACTTTCAAATAGTTAAATAAATAGGGAAATAGATTGACAGTTAATACGAATATTAAGAGAGAGTGCGTTAGCTAGATAACTTTAGGAAGAACATTATATCCTTTTTGAGCCAACAAATGTTTATAAAAATTTAAATAATAATACAAGGCAGAATAATAGAGATTAGATTATATCATAATTCTAATCTTTGTAGAATTGTTTGGTGTACTAGCATTCATAACATTAAACAAAATAACCTTTAAGAACTGTATTTAATTTTTAGTTATTTAATTCCATTTAGGCCATGAGTCTTTCAAGAAAAAGTCCTATTTGGGAATTCTTCACATCTTGTGAGGATGAATTAAAAGCAGAATGTCAACTTTGCCACCTCAAAATTTCACGAGGAGGAAAAACTCCAAAATCATGTACAACAACATCCATGATAAATCATCTTCGATCCAGACATAATAAAGAATATAAAGAGTATGAAGAAAAGAAGAAAGTGTTAGAGAAAAATGAGGTACCCAAAACACATTTTGTTCTCACTAATCAAACTTCAAAACAACAGACTTTACAAGAATGCTTTAAGAAGACAATTCCTTATAGTTTTGAGTCTATGGAGGCTAAAAATTTAAACAAAAAGCTTGTCGAAATGATTGCTATAGACTGCCAACCATTTTCAGTAGTTGAAGACGAAGGATTTTTGAGATTTGCCCAAGCTATGAATCCTAGGTACAAAGTTCCTTCTAGAAAACATGTTACTGAAAAACTTTTTCCTGAATTATATTCTGCTGTGCACAAAAAGGTTAGTGATATTATTAAATGTGCTACTTCTGTATCTTGTACATCAGACATTTGGACATGCAGAAACAACCAGAAGTCTTTTTTAAGTTTGACAGCACGGTGGATTAATGATGAATTCAACTATGCTTCTGCTTTACTTGCTTGCCGAGAGATTTCTGGGTCTCACACAGCTAACAACACAGTGAATCTGACTGAAGAAATATTTATAGAATGGAAAATCAGTAAAGAAAAGTGTCATGCAATTGTCATTGACAATGGAACAAACATGGTGAAGGCTATGAGTGATTTAGGAATCACAAGCATACGATGTTTTGTTCACACCCTTCAGTTGTGTGTTCAGAAAGTAATTATAGCACAGCGTAATGTAACAGAATTACTGGCCGTAAGCAGAAAGATAGTGGGCCACTTTCATCATTCTTCTTCAGCCAAAGACAAACTACATGACATACAAGAGATTTTGGAACTGCCTTGTCATGTACTTATACAAGATGTTCAAACAAGATGGAATAGCAGCTATTACATGTTAAAAAGGCTGCTTGAACAAAAGAAAGCCTTAATACTGTACTTTGCAGATAACAATACCAACATAGCAGGACTTACTAACAACCAATGGTTGCTTGCAGAGAAAGTAGTTCGTTTGCTGGAACCTGCTGAAGAAGTAACCAAACTGATGTGCCTTGAGAGTGCAAGTATTTCTTACATGATACCTGCTGTAAAGATCCTCCTATCATTCTACTCCAGGGAAGATGCTGAAATTGGTGTGCATGCAATGAAAGCTGAGATGCAGATGGAAATTCAGAGACGATTTTGTTATCTTGAAAGCCAATTACATTGCTCCATTGCCACATTTCTAGATCCAAGGTTTAAGGACAAATTTTTCTCAAGTGATTGTGTGAAGCAGCAAGTGCATGCAAAAATATTAGAAATAAAANATATAAGTGAGGGCAACAATCTTAAGTCTGCTTCTGAAAATATTCAGAAAGCATCTTCTTCAAGCAGTGCGCAAAGCTTCAGTTTGTGGGATTGCTTCAATGAAATTGCATCATCACCATCGACCAAATCTTCAAGTCCAGGAGTTGAAGAGGAGATAAGTCTTTACCTTAGTCGGTTACTATTAGAGAGGAAGGAAAGCTTAATAACTTGGTGGCAACATCACCATCAGCAGTACCCTATACTGGCAGAGCTAGCCAAAAAATATCTTGCTCCTCCACCGACCTCAGTTGAAAGTGAGCGTCTATTTAGTATCGCAAATGTGCAGTATAGTGACCGCAGGAGCAATCTAGCAGCTGACAGTGCTGAGAAATTATTGTTCCTGCACTATAATGTCAAACTTTAAAATTTGACTACTAAAATTTTTAATAATAAAATTGTAATAATAATTAANTTTTGANATAAATTTCAGATTTATTCTTGTATCCCAGAAGTTCATTCTATGATGGTGATATTCGGCTTCGGCCGAATATTGATTTTAACACTCGGCCATTCGGCATTCGGCCAAAATGTGTATTCGGTGCACCTCTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| EuthAT-2 | TBX21 | 1541 | 1550 | + | 15.40 | TTCACACCCT |
| EuthAT-2 | ERF057 | 2428 | 2441 | + | 15.33 | GCTCCTCCACCGAC |
| EuthAT-2 | crc | 2263 | 2273 | - | 15.07 | GATGATGCAAT |
| EuthAT-2 | SoxN | 2256 | 2266 | - | 15.07 | CAATTTCATTG |
| EuthAT-2 | ERF012 | 2434 | 2441 | + | 15.03 | CCACCGAC |
| EuthAT-2 | PI | 912 | 925 | + | 15.02 | AGAAAAGAAGAAAG |
| EuthAT-2 | Tbx6 | 1541 | 1549 | - | 14.98 | GGGTGTGAA |
| EuthAT-2 | RAP2-9 | 2435 | 2443 | + | 14.92 | CACCGACCT |
| EuthAT-2 | HDG1 | 388 | 397 | + | 14.91 | TAATTAATGA |
| EuthAT-2 | ZNF317 | 1210 | 1217 | - | 14.80 | ACAGCAGA |
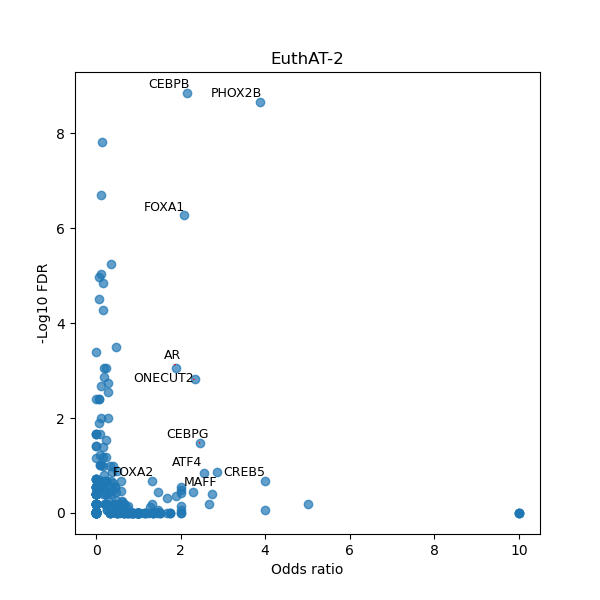
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.