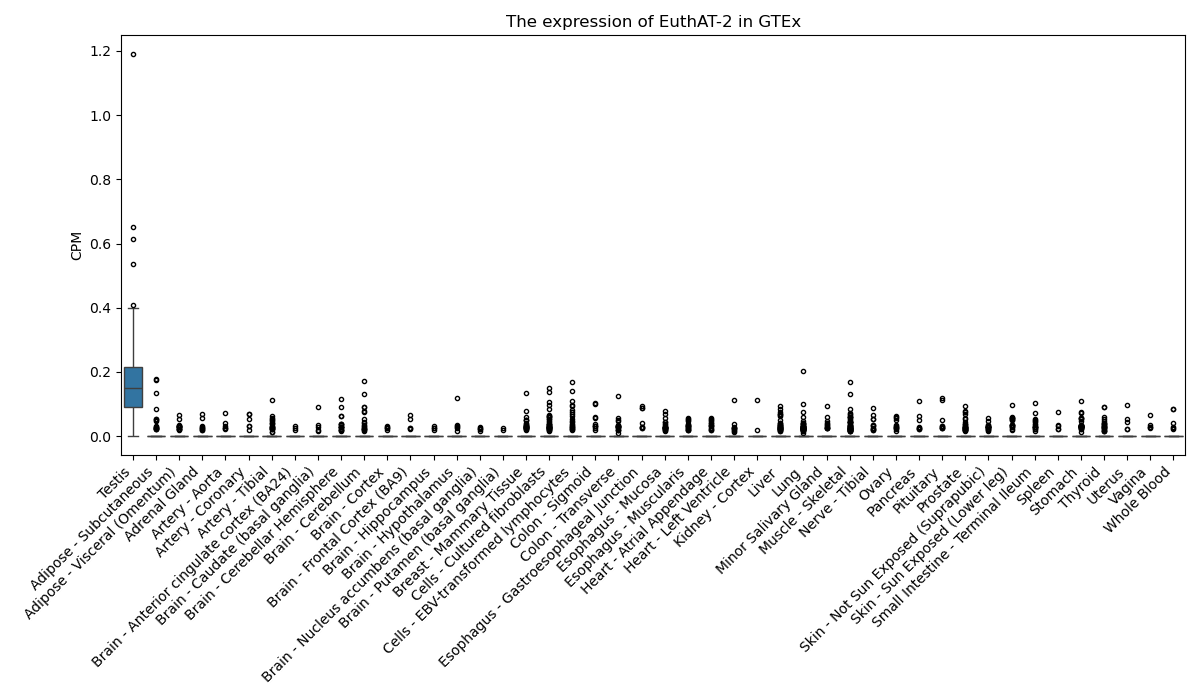
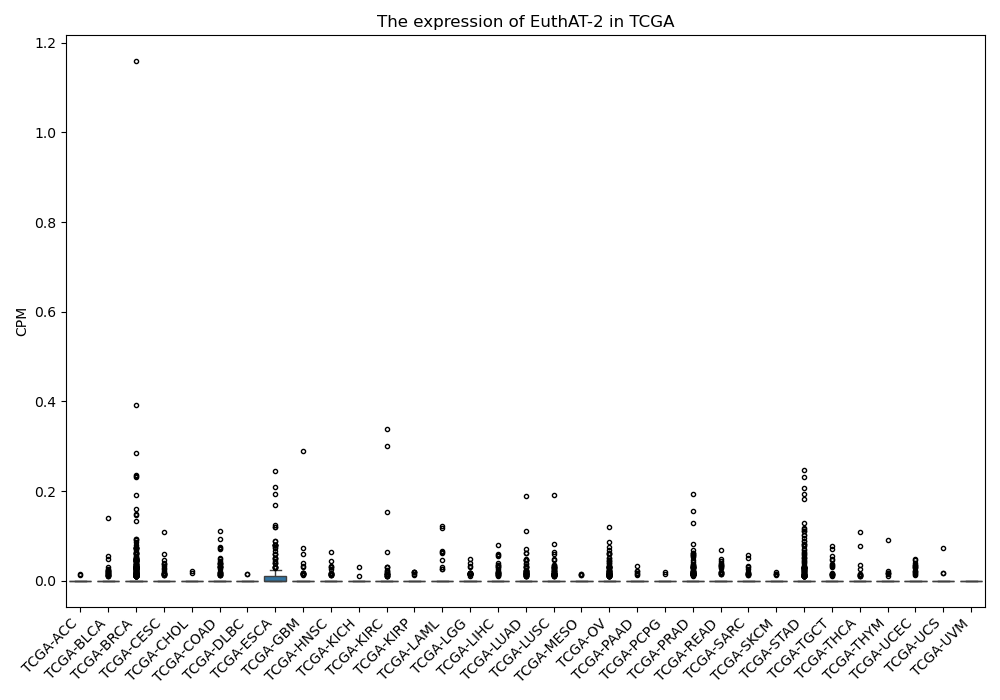
EuthAT-2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001172 |
|---|---|
| TE superfamily | Activator |
| TE class | DNA |
| Species | Eutheria |
| Length | 2747 |
| Kimura value | 25.50 |
| Tau index | 1.0000 |
| Description | Ancient hAT DNA transposon from eutherian mammals. |
| Comment | >74% identical to consensus. It encodes remnants of transposase. No copies in opossum. ORF 727-2562 encoded a transposase closest related to HAT-12_XT_tp of Xenopus and HAT5_CI_tp and HAT1_CI_tp of Ciona. Present at orthologous sites throughout Eutheria, but not in marsupials. |
| Sequence |
TAGGGATGCACCGAAGTTCGGCTTCGGCCTCGGTATTCGGCCAAAATCTGCTATTCGGCTTCGGCTTCGGCCTCGGCCTTGGAAGGCCGAATAGCCGAATGCCTACTTCAACGTCATTTCTGGCATCCCTGAACTTGTGCGTGATCAATGATCGTGTGATCGCGTAGGTTACCATCATATCTAATCAGCGTAACAACGTCAGTAATACGAACAAGGCATGTCATTCGGATTTATTGTTAATATCTGAATATTCACTTTCAAATATAGGTAAGCATTCTAGATGTAACTTGGAGTGAAAAATCTCCTTATGTGCTTTATATGTTGAAATTCTATCAGTCAGTTGAAGTATAAATGTATATTTATTATTGCAAGAGTAAAATGCTGCAATAATTAATGATGATTGTGAAAGCTGTTAATTTTATGTACAAAAACAATTATTAAGAACTTTCAAATAGTTAAATAAATAGGGAAATAGATTGACAGTTAATACGAATATTAAGAGAGAGTGCGTTAGCTAGATAACTTTAGGAAGAACATTATATCCTTTTTGAGCCAACAAATGTTTATAAAAATTTAAATAATAATACAAGGCAGAATAATAGAGATTAGATTATATCATAATTCTAATCTTTGTAGAATTGTTTGGTGTACTAGCATTCATAACATTAAACAAAATAACCTTTAAGAACTGTATTTAATTTTTAGTTATTTAATTCCATTTAGGCCATGAGTCTTTCAAGAAAAAGTCCTATTTGGGAATTCTTCACATCTTGTGAGGATGAATTAAAAGCAGAATGTCAACTTTGCCACCTCAAAATTTCACGAGGAGGAAAAACTCCAAAATCATGTACAACAACATCCATGATAAATCATCTTCGATCCAGACATAATAAAGAATATAAAGAGTATGAAGAAAAGAAGAAAGTGTTAGAGAAAAATGAGGTACCCAAAACACATTTTGTTCTCACTAATCAAACTTCAAAACAACAGACTTTACAAGAATGCTTTAAGAAGACAATTCCTTATAGTTTTGAGTCTATGGAGGCTAAAAATTTAAACAAAAAGCTTGTCGAAATGATTGCTATAGACTGCCAACCATTTTCAGTAGTTGAAGACGAAGGATTTTTGAGATTTGCCCAAGCTATGAATCCTAGGTACAAAGTTCCTTCTAGAAAACATGTTACTGAAAAACTTTTTCCTGAATTATATTCTGCTGTGCACAAAAAGGTTAGTGATATTATTAAATGTGCTACTTCTGTATCTTGTACATCAGACATTTGGACATGCAGAAACAACCAGAAGTCTTTTTTAAGTTTGACAGCACGGTGGATTAATGATGAATTCAACTATGCTTCTGCTTTACTTGCTTGCCGAGAGATTTCTGGGTCTCACACAGCTAACAACACAGTGAATCTGACTGAAGAAATATTTATAGAATGGAAAATCAGTAAAGAAAAGTGTCATGCAATTGTCATTGACAATGGAACAAACATGGTGAAGGCTATGAGTGATTTAGGAATCACAAGCATACGATGTTTTGTTCACACCCTTCAGTTGTGTGTTCAGAAAGTAATTATAGCACAGCGTAATGTAACAGAATTACTGGCCGTAAGCAGAAAGATAGTGGGCCACTTTCATCATTCTTCTTCAGCCAAAGACAAACTACATGACATACAAGAGATTTTGGAACTGCCTTGTCATGTACTTATACAAGATGTTCAAACAAGATGGAATAGCAGCTATTACATGTTAAAAAGGCTGCTTGAACAAAAGAAAGCCTTAATACTGTACTTTGCAGATAACAATACCAACATAGCAGGACTTACTAACAACCAATGGTTGCTTGCAGAGAAAGTAGTTCGTTTGCTGGAACCTGCTGAAGAAGTAACCAAACTGATGTGCCTTGAGAGTGCAAGTATTTCTTACATGATACCTGCTGTAAAGATCCTCCTATCATTCTACTCCAGGGAAGATGCTGAAATTGGTGTGCATGCAATGAAAGCTGAGATGCAGATGGAAATTCAGAGACGATTTTGTTATCTTGAAAGCCAATTACATTGCTCCATTGCCACATTTCTAGATCCAAGGTTTAAGGACAAATTTTTCTCAAGTGATTGTGTGAAGCAGCAAGTGCATGCAAAAATATTAGAAATAAAANATATAAGTGAGGGCAACAATCTTAAGTCTGCTTCTGAAAATATTCAGAAAGCATCTTCTTCAAGCAGTGCGCAAAGCTTCAGTTTGTGGGATTGCTTCAATGAAATTGCATCATCACCATCGACCAAATCTTCAAGTCCAGGAGTTGAAGAGGAGATAAGTCTTTACCTTAGTCGGTTACTATTAGAGAGGAAGGAAAGCTTAATAACTTGGTGGCAACATCACCATCAGCAGTACCCTATACTGGCAGAGCTAGCCAAAAAATATCTTGCTCCTCCACCGACCTCAGTTGAAAGTGAGCGTCTATTTAGTATCGCAAATGTGCAGTATAGTGACCGCAGGAGCAATCTAGCAGCTGACAGTGCTGAGAAATTATTGTTCCTGCACTATAATGTCAAACTTTAAAATTTGACTACTAAAATTTTTAATAATAAAATTGTAATAATAATTAANTTTTGANATAAATTTCAGATTTATTCTTGTATCCCAGAAGTTCATTCTATGATGGTGATATTCGGCTTCGGCCGAATATTGATTTTAACACTCGGCCATTCGGCATTCGGCCAAAATGTGTATTCGGTGCACCTCTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| EuthAT-2 | ONECUT3 | 2686 | 2697 | - | 15.65 | TAAAATCAATAT |
| EuthAT-2 | Trl | 498 | 506 | + | 15.63 | AAGAGAGAG |
| EuthAT-2 | RVE1 | 2418 | 2426 | + | 15.59 | AAAATATCT |
| EuthAT-2 | Pgr | 950 | 966 | - | 15.54 | GAGAACAAAATGTGTTT |
| EuthAT-2 | NAC105 | 1838 | 1846 | - | 15.51 | CAAGCAACC |
| EuthAT-2 | ONECUT1 | 2687 | 2695 | - | 15.50 | AAATCAATA |
| EuthAT-2 | ZBTB32 | 1783 | 1792 | - | 15.47 | AGTACAGTAT |
| EuthAT-2 | RVE7L | 2418 | 2426 | + | 15.47 | AAAATATCT |
| EuthAT-2 | ERF055 | 2434 | 2442 | - | 15.42 | GGTCGGTGG |
| EuthAT-2 | Pgr | 530 | 546 | - | 15.42 | AAGGATATAATGTTCTT |
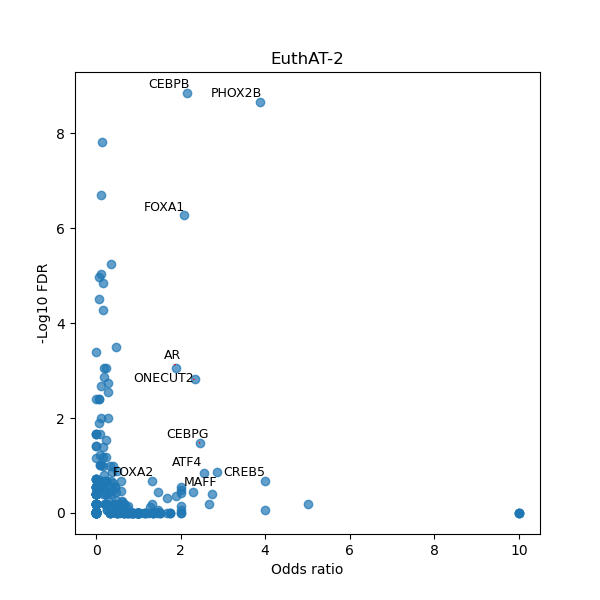
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.