Eulor9C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000141 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 270 |
| Kimura value | 25.08 |
| Tau index | 0.0000 |
| Description | Eulor9C (Euteleostomi-conserved low frequency repeat 9C) |
| Comment | Putative DNA transposon (assignment unclear). Present in mammals and birds (>100 copies). Entirety of Eulor9C forms one long, imperfect hairpin. |
| Sequence |
TATGAATATTAAATACAAAAAACCGCGTTNAAATAACGTTAAGGGTCTGACTTAAACCCACAAAAGCNAGGAAATTCAGAGTTAAGGCTGACACTCCGTCCTTAACTCACCCCGTCGTGCCTGGGTATCTNTTNATTTTCTTTNNNATAGGCACAACGGCGTGAGTTAAGGACGGAGTGTCAGCCTTAACTTTGAATTTCCTGGCTTTTGTCGGTTTGTGTCACAACCTTAACGTTATTTAAACGTAGTTTTTTGTATTTAATATTCATA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor9C | PHYPA_011264 | 209 | 216 | - | 12.39 | AACCGACA |
| Eulor9C | PHYPADRAFT_28324 | 209 | 216 | + | 12.38 | TGTCGGTT |
| Eulor9C | pan | 13 | 26 | - | 12.34 | GCGGTTTTTTGTAT |
| Eulor9C | ERF027 | 209 | 215 | - | 12.31 | ACCGACA |
| Eulor9C | PHYPADRAFT_64121 | 209 | 216 | + | 12.27 | TGTCGGTT |
| Eulor9C | DREB2E | 207 | 216 | - | 12.02 | AACCGACAAA |
| Eulor9C | PK21687.1 | 53 | 59 | + | 11.89 | TAAACCC |
| Eulor9C | NLP7 | 202 | 209 | + | 11.82 | TGGCTTTT |
| Eulor9C | DREB2C | 209 | 216 | - | 11.79 | AACCGACA |
| Eulor9C | ARF5 | 209 | 215 | - | 11.79 | ACCGACA |
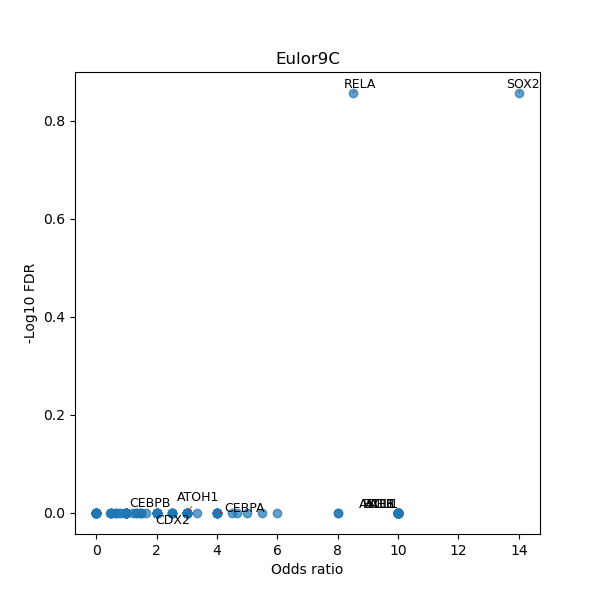
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.