Eulor9B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000140 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 240 |
| Kimura value | 23.10 |
| Tau index | 0.0000 |
| Description | Eulor9B (Euteleostomi-conserved low frequency repeat 9B) |
| Comment | Putative DNA transposon (assignment unclear). Present in mammals and birds (~40 copies). Has a hairpin-tail structure typical for all Eulor repeats. However, termini are poorly defined. Eulor9B is slightly more similar to the reverse complement of Eulor9A. |
| Sequence |
CAATATGTGATGGCAACATTACGGTTGAGATTTTAAACGCACAAAATGTCAGGAAATTCAAAGTTATGGTTCCCACGGCAACCGTAACTCGGCCACATTGCACATATATATTAAGGCATTAAAGTTAACACCATNCGCAGTCACGNAANAAACTATATATGCATAAGGGGGCGGAGTTACGGTTGCTATGGGAACCATAACTTTGAATTTCCTGACTTTTGTGCGTTTGAATTTNCTNGC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor9B | unc-86 | 157 | 164 | + | 14.03 | ATATGCAT |
| Eulor9B | KLF12 | 168 | 176 | + | 13.96 | GGGGCGGAG |
| Eulor9B | KLF14 | 168 | 176 | + | 13.93 | GGGGCGGAG |
| Eulor9B | nhr-6 | 44 | 51 | + | 13.81 | AAATGTCA |
| Eulor9B | KLF10 | 168 | 176 | + | 13.80 | GGGGCGGAG |
| Eulor9B | TCX3 | 199 | 210 | - | 13.77 | AAATTCAAAGTT |
| Eulor9B | TCX3 | 54 | 65 | + | 13.77 | AAATTCAAAGTT |
| Eulor9B | ZNF281 | 167 | 176 | + | 13.57 | GGGGGCGGAG |
| Eulor9B | TCP7 | 66 | 76 | - | 13.56 | GTGGGAACCAT |
| Eulor9B | RFX2 | 69 | 82 | - | 13.53 | GTTGCCGTGGGAAC |
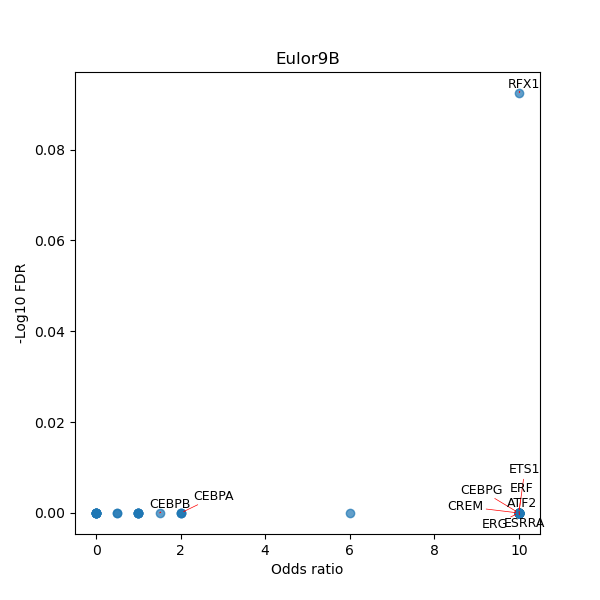
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.