Eulor9B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000140 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 240 |
| Kimura value | 23.10 |
| Tau index | 0.0000 |
| Description | Eulor9B (Euteleostomi-conserved low frequency repeat 9B) |
| Comment | Putative DNA transposon (assignment unclear). Present in mammals and birds (~40 copies). Has a hairpin-tail structure typical for all Eulor repeats. However, termini are poorly defined. Eulor9B is slightly more similar to the reverse complement of Eulor9A. |
| Sequence |
CAATATGTGATGGCAACATTACGGTTGAGATTTTAAACGCACAAAATGTCAGGAAATTCAAAGTTATGGTTCCCACGGCAACCGTAACTCGGCCACATTGCACATATATATTAAGGCATTAAAGTTAACACCATNCGCAGTCACGNAANAAACTATATATGCATAAGGGGGCGGAGTTACGGTTGCTATGGGAACCATAACTTTGAATTTCCTGACTTTTGTGCGTTTGAATTTNCTNGC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor9B | TCP21 | 66 | 76 | + | 9.21 | ATGGTTCCCAC |
| Eulor9B | TSO1 | 23 | 37 | + | 9.11 | GGTTGAGATTTTAAA |
| Eulor9B | TCP9 | 66 | 76 | + | 9.07 | ATGGTTCCCAC |
| Eulor9B | Su(H) | 187 | 196 | + | 8.68 | TATGGGAACC |
| Eulor9B | TSO1 | 202 | 216 | - | 8.61 | GTCAGGAAATTCAAA |
| Eulor9B | TSO1 | 48 | 62 | + | 8.61 | GTCAGGAAATTCAAA |
| Eulor9B | EGR3 | 164 | 174 | - | 6.19 | CCGCCCCCTTA |
| Eulor9B | THRB | 198 | 215 | + | 4.00 | TAACTTTGAATTTCCTGA |
| Eulor9B | THRB | 49 | 66 | - | 4.00 | TAACTTTGAATTTCCTGA |
| Eulor9B | ZSCAN4 | 38 | 52 | + | 1.88 | CGCACAAAATGTCAG |
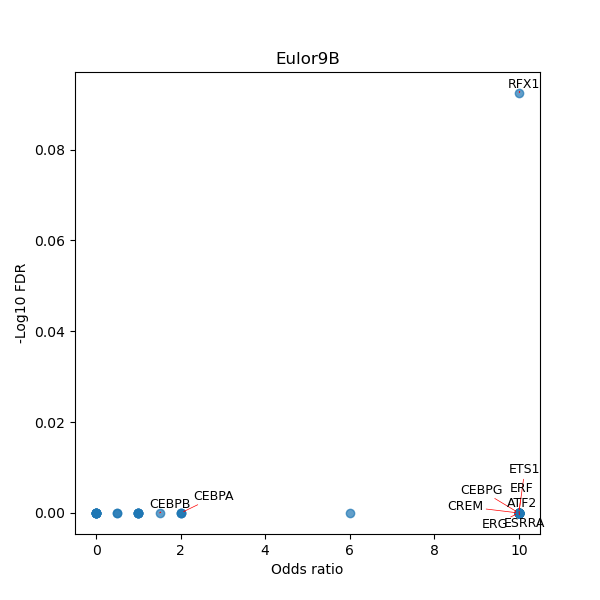
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.