Eulor9A
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000139 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 301 |
| Kimura value | 26.20 |
| Tau index | 0.0000 |
| Description | Eulor9A (Euteleostomi-conserved low frequency repeat 9A) |
| Comment | Putative DNA transposon (assignment unclear). Present in mammals and birds (>100 copies). Has a hairpin-tail structure typical for all Eulor repeats. However, termini are poorly defined. |
| Sequence |
TAAATTAAATGCAAAAAACTACGTTAATGCAACGTTATGGCTGAGAATTTAAACGCACAAAAGTCAGGAAATTCAAAGTTACGGTTCCCACAGCAACCGTAACTCGGCCCCCTTGCGCATAGNAGTATTTTGTATTACTGTATATGNTGTTAAATTTATGCCTTAATATCAGTATGTGCAATGTGGCCGAGTTACGGTTGCTGTGGGAACCGTAACTTTGAATTTCCTGACTTTTGTGCGTTTAAAATCTCAACCGTAACGTTGCATTAACGTAGGNTTTTTTGCATTTTACTTTCCTTTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor9A | RFX5 | 84 | 97 | + | 13.69 | GTTCCCACAGCAAC |
| Eulor9A | RFX5 | 197 | 210 | + | 13.53 | GTTGCTGTGGGAAC |
| Eulor9A | RFX5 | 84 | 97 | - | 13.53 | GTTGCTGTGGGAAC |
| Eulor9A | DOF5.8 | 279 | 297 | + | 13.41 | TTTTTGCATTTTACTTTCC |
| Eulor9A | rib | 10 | 18 | + | 13.33 | TGCAAAAAA |
| Eulor9A | rib | 278 | 286 | - | 13.33 | TGCAAAAAA |
| Eulor9A | ZBTB32 | 135 | 144 | - | 13.15 | TATACAGTAA |
| Eulor9A | MYB98 | 256 | 263 | - | 12.95 | AACGTTAC |
| Eulor9A | M1BP | 179 | 189 | - | 12.70 | CGGCCACATTG |
| Eulor9A | tbx-37 | 38 | 49 | + | 12.49 | TGGCTGAGAATT |
TFBS enrichment in GRCh38
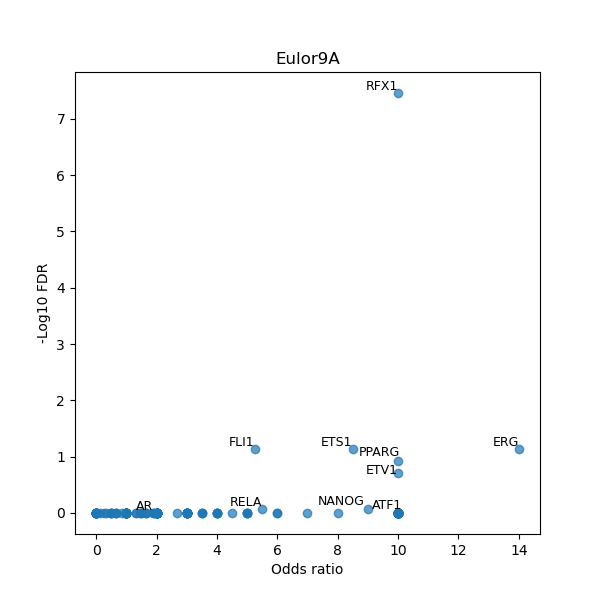
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.