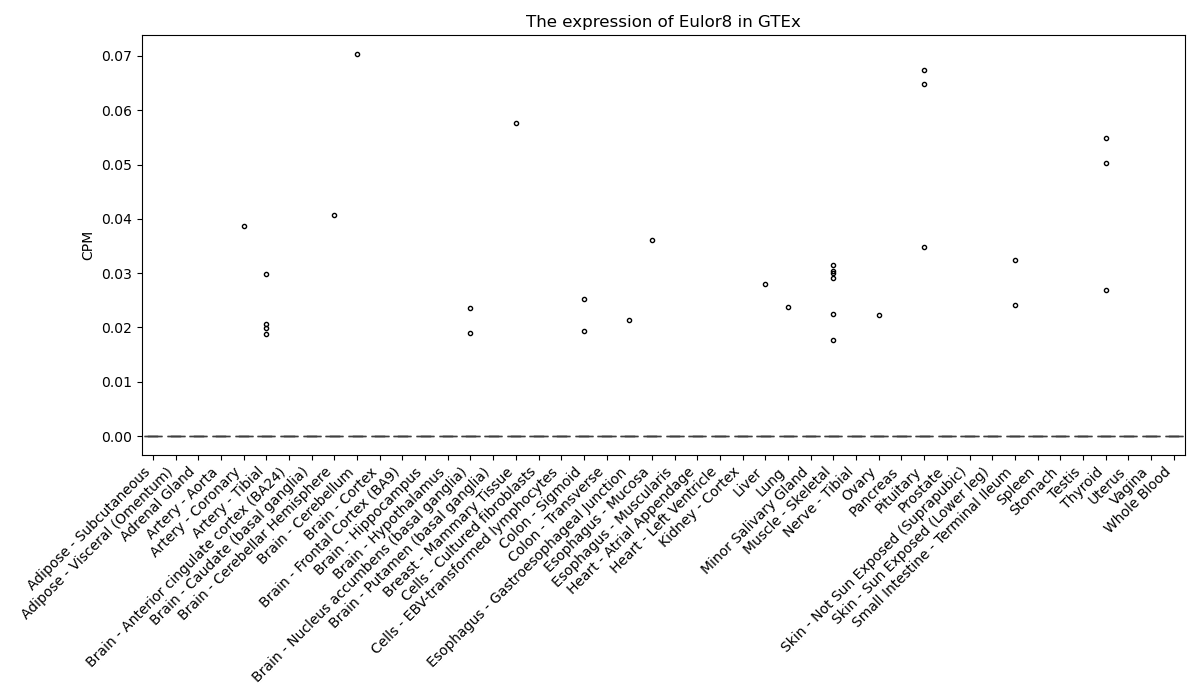
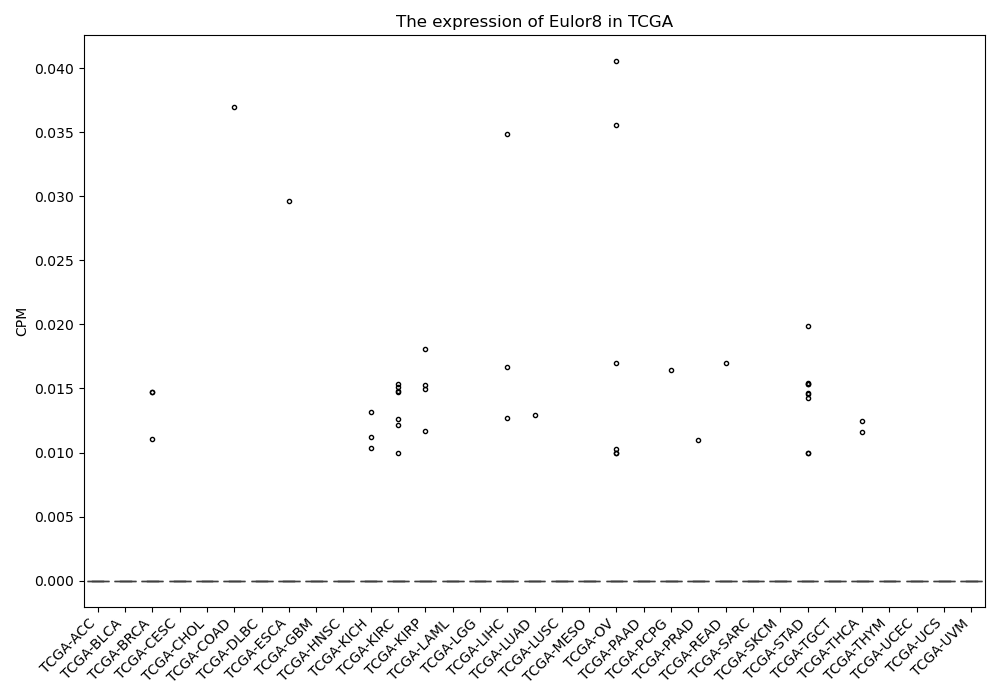
Eulor8
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000138 |
|---|---|
| TE superfamily | Tc1-Mariner |
| TE class | DNA |
| Species | Amniota |
| Length | 562 |
| Kimura value | 30.49 |
| Tau index | 0.0000 |
| Description | Eulor8 (Euteleostomi-conserved low frequency repeat 8) |
| Comment | Putative DNA transposon (assignment unclear). Eulor8 forms an imperfect 562 bp hairpin, with no tail. Element boundary unclear. Ends unsure, though CAG...CTG make for familiar termini flanked by TA TSDs. |
| Sequence |
CAGCATCCGAGGAGANNCATAAGATTNCAATCTCGAGATTGTAACGAGAATGGGTTAAGTTTGTACCATTTCCCATTCTCGTTACAATCNTCGTCACGAGAATGGATTCATGTTGTGCCATTTTCTGTTCTCGTTACAATCTTTGTCACGAGAACGAGGTATCAGAATTTTAATGCCTAATACGTTCTGTGAAATACGGCAGCGTGCTNTACTGCTTTGACCTTTTCAATATTCTGCATTTTGATTGGCTGGCCATTCCGCCTCTTCCTCACAGGTTANCGAGGCTGTAAACAGGGGAGAACGGGAATGGCCAGCCAATCAAATTACAGAATATTGAAAAGGTCAAAGTAGTACAGCATACTGCTATAATTCACAGAATATATGAGGCATTAAAATTCCGATACCTCATTCTCGTGACAAAGATTGTAACGAGAACAGAAAATGGCACGACATGAATCCGTTCTCGTGACAANGATTGTAACGAGAACGGGAAATGGTACAAANTTAACCCATTCTCGTTACAATCTTAAGATTGTAATCTCGTGGTCCTCCTCAGATGCCG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor8 | TCF7L1 | 338 | 349 | + | 13.76 | AAAGGTCAAAGT |
| Eulor8 | WRKY33 | 215 | 224 | - | 13.68 | AAGGTCAAAG |
| Eulor8 | WRKY33 | 339 | 348 | + | 13.68 | AAGGTCAAAG |
| Eulor8 | WRKY46 | 215 | 224 | + | 13.64 | CTTTGACCTT |
| Eulor8 | WRKY46 | 339 | 348 | - | 13.64 | CTTTGACCTT |
| Eulor8 | sex-1 | 217 | 224 | - | 13.55 | AAGGTCAA |
| Eulor8 | sex-1 | 339 | 346 | + | 13.55 | AAGGTCAA |
| Eulor8 | ERR | 218 | 225 | - | 13.48 | AAAGGTCA |
| Eulor8 | ERR | 338 | 345 | + | 13.48 | AAAGGTCA |
| Eulor8 | PHOX2B | 316 | 327 | + | 13.44 | CAATCAAATTAC |
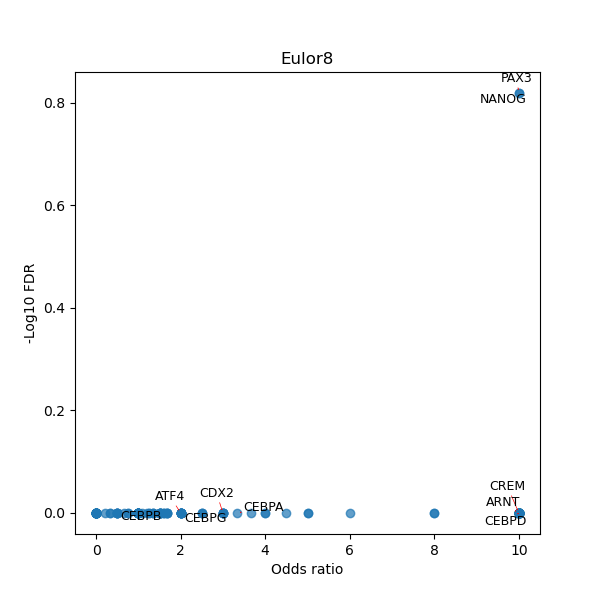
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.