Eulor7
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000137 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 193 |
| Kimura value | 27.28 |
| Tau index | 0.0000 |
| Description | Eulor7 (Euteleostomi-conserved low frequency repeat 7) |
| Comment | Putative DNA transposon (assignment unclear). Fewer than 20 copies in human, 40-50 copies in alligator. Position (roughly) 38-175 is an imperfect hairpin. |
| Sequence |
TTCATAATATATGGATGGTAGAATTCTACCATTGTTAAGCTGTCAGTCAAAAACCTTGTTATACATGGATGTGACGTCANATCCATGTATAACAAACTTTCCTGTCAATCAAAAATCTTGTTATACATGGATNTGACGTCAGATCCATGTATAACAAGGTTTTTGACTGACAGCTNAGCNGCGGAACACGGCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor7 | Sox7 | 29 | 38 | - | 10.94 | TTAACAATGG |
| Eulor7 | ceh-60 | 164 | 172 | + | 10.56 | TGACTGACA |
| Eulor7 | ceh-60 | 41 | 49 | - | 10.56 | TGACTGACA |
| Eulor7 | FLI1::FOXI1 | 98 | 108 | - | 10.53 | TTGACAGGAAA |
| Eulor7 | SOX21 | 22 | 36 | - | 7.31 | AACAATGGTAGAATT |
| Eulor7 | Sox21b | 22 | 36 | + | -0.89 | AATTCTACCATTGTT |
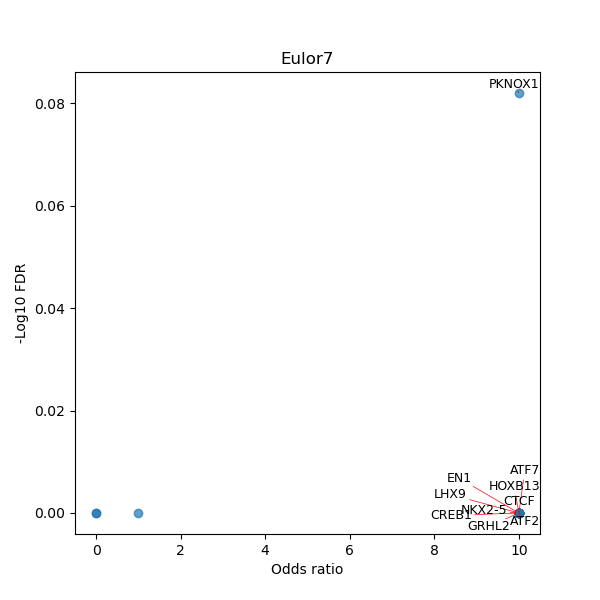
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.