Eulor7
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000137 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 193 |
| Kimura value | 27.28 |
| Tau index | 0.0000 |
| Description | Eulor7 (Euteleostomi-conserved low frequency repeat 7) |
| Comment | Putative DNA transposon (assignment unclear). Fewer than 20 copies in human, 40-50 copies in alligator. Position (roughly) 38-175 is an imperfect hairpin. |
| Sequence |
TTCATAATATATGGATGGTAGAATTCTACCATTGTTAAGCTGTCAGTCAAAAACCTTGTTATACATGGATGTGACGTCANATCCATGTATAACAAACTTTCCTGTCAATCAAAAATCTTGTTATACATGGATNTGACGTCAGATCCATGTATAACAAGGTTTTTGACTGACAGCTNAGCNGCGGAACACGGCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor7 | atf-7 | 71 | 79 | + | 14.38 | GTGACGTCA |
| Eulor7 | BZIP60 | 134 | 141 | + | 14.25 | TGACGTCA |
| Eulor7 | BZIP60 | 134 | 141 | - | 14.25 | TGACGTCA |
| Eulor7 | BZIP60 | 72 | 79 | + | 14.25 | TGACGTCA |
| Eulor7 | BZIP60 | 72 | 79 | - | 14.25 | TGACGTCA |
| Eulor7 | BZIP52 | 168 | 175 | + | 13.59 | TGACAGCT |
| Eulor7 | BZIP52 | 38 | 45 | - | 13.59 | TGACAGCT |
| Eulor7 | MEIS2 | 101 | 108 | - | 13.53 | TTGACAGG |
| Eulor7 | CREB3L4 | 134 | 142 | - | 13.53 | CTGACGTCA |
| Eulor7 | NAC035 | 187 | 193 | + | 13.30 | CACGGCA |
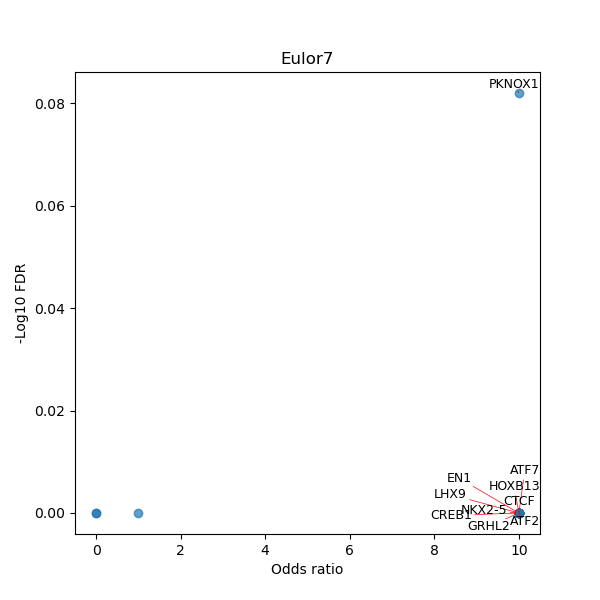
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.