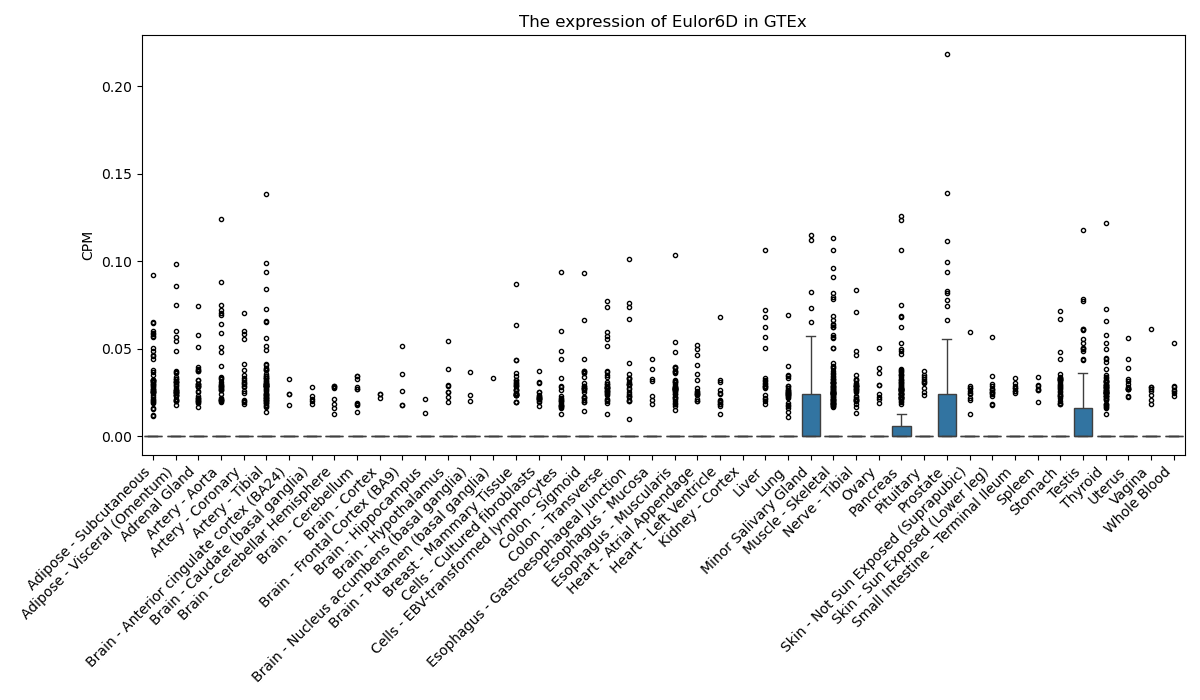
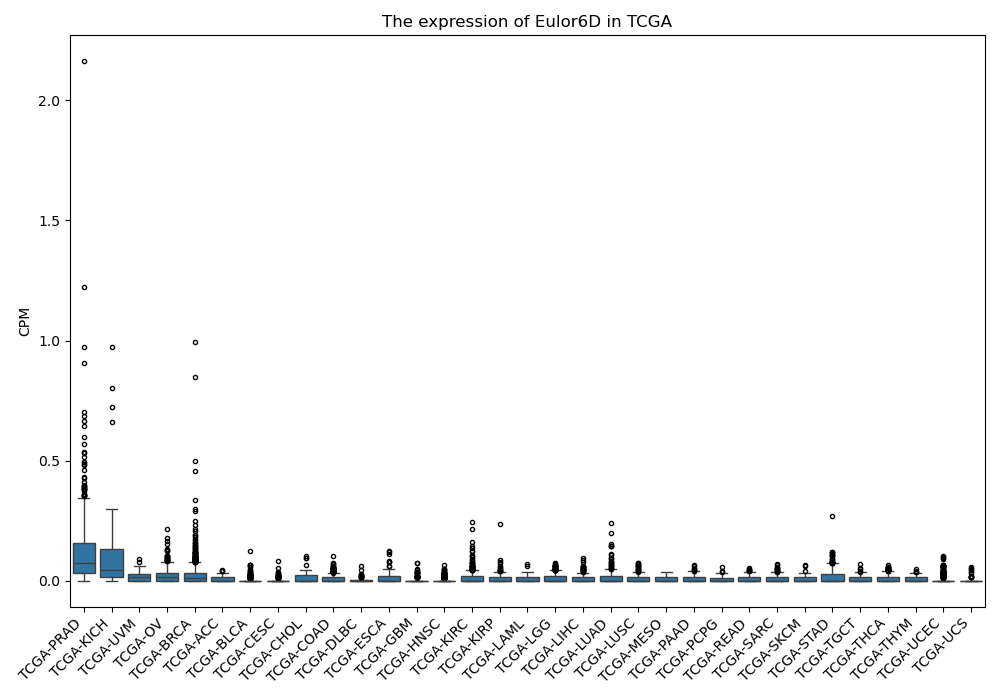
Eulor6D
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000135 |
|---|---|
| TE superfamily | Crypton |
| TE class | DNA |
| Species | Tetrapoda |
| Length | 302 |
| Kimura value | 28.95 |
| Tau index | 0.0000 |
| Description | Eulor6D (Euteleostomi-conserved low frequency repeat 6D) |
| Comment | Putative DNA transposon (assignment unclear). Present in ~100 copies in the human genome, ~100 in platypus, also detected in Xenopus. Position (roughly) 1-162 is an imperfect hairpin. |
| Sequence |
TAATTAAGCAATAAGACACGACAGGCAGTGCATTTCTGGGCGATTATAGCACGCCTCGGGTGGCGTTATAAGGCACGAGGCCGAAGGCCGAGTGACTTTAACCACCCGAGAAGTGCAATAATCGCCCNGAAATGCACTGCCTGGAGTGTCTTATTGCTATTATGAAATGGAATTTATACATAAAAATAAGGAAAACAGTCAGACCCGTGCATTTACTGGGCATTATTGACATGGGCGTGACATCACCGACAGCCAATCAGAAANCTCCGNTTGCGTCCGGNGTTCTAAAGCCGTTTCATAAT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor6D | ERF036 | 244 | 251 | - | 15.11 | TGTCGGTG |
| Eulor6D | HAP3 | 251 | 259 | - | 15.11 | TGATTGGCT |
| Eulor6D | ERF010 | 244 | 251 | + | 15.07 | CACCGACA |
| Eulor6D | TINY | 243 | 251 | + | 14.92 | TCACCGACA |
| Eulor6D | DREB2C | 244 | 251 | + | 14.85 | CACCGACA |
| Eulor6D | PHYPADRAFT_173530 | 244 | 251 | - | 14.79 | TGTCGGTG |
| Eulor6D | cebp-1 | 158 | 168 | - | 14.76 | ATTTCATAATA |
| Eulor6D | PHYPA_011264 | 244 | 251 | + | 14.67 | CACCGACA |
| Eulor6D | PHYPADRAFT_28324 | 244 | 251 | - | 14.66 | TGTCGGTG |
| Eulor6D | NFYA | 253 | 260 | + | 14.54 | CCAATCAG |
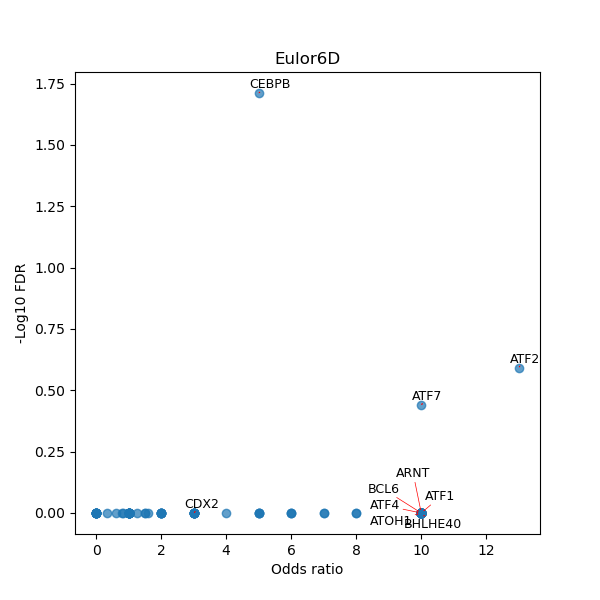
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.