Eulor6C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000134 |
|---|---|
| TE superfamily | Crypton |
| TE class | DNA |
| Species | Tetrapoda |
| Length | 208 |
| Kimura value | 24.05 |
| Tau index | 0.0000 |
| Description | Eulor6C (Euteleostomi-conserved low frequency repeat 6C) |
| Comment | Putative DNA transposon (assignment unclear). Position (roughly) 1-160 is an imperfect hairpin. |
| Sequence |
TTAATATGGCATTAAGACACGACAGGGCGTGTTTTANTGGTCCATTAATACATGCCTCGGGTGCGTTGCGAGGCACAAGGCTNNAGGCCAAGTACTTCGACCACTCGAGGCATATATTAATGGNCCAATAAAGCACGCCCCGGAGTGTCTTAATACTANCANAATACGGCTCTTTAATTTCCTATTNAATTTTNAAGGNTTCTTTTTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor6C | HOXB13 | 125 | 133 | + | 10.68 | CCAATAAAG |
| Eulor6C | bsh | 117 | 123 | + | 10.57 | TTAATGG |
| Eulor6C | bsh | 42 | 48 | - | 10.57 | TTAATGG |
| Eulor6C | ZIC4 | 134 | 147 | + | 10.35 | CACGCCCCGGAGTG |
| Eulor6C | ZBED4 | 134 | 143 | + | 10.17 | CACGCCCCGG |
| Eulor6C | SP2 | 133 | 141 | - | 10.06 | GGGGCGTGC |
| Eulor6C | SP3 | 132 | 142 | + | 9.56 | AGCACGCCCCG |
| Eulor6C | CG4328 | 126 | 132 | - | 9.51 | TTTATTG |
| Eulor6C | TP53 | 16 | 33 | - | 9.27 | AACACGCCCTGTCGTGTC |
| Eulor6C | ZIC1 | 18 | 31 | - | 8.34 | CACGCCCTGTCGTG |
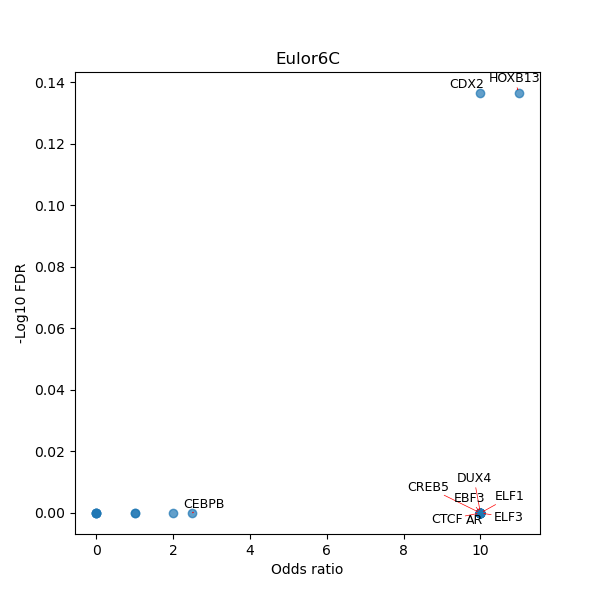
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.