Eulor5B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000131 |
|---|---|
| TE superfamily | Crypton-A |
| TE class | DNA |
| Species | Tetrapoda |
| Length | 192 |
| Kimura value | 22.48 |
| Tau index | 0.0000 |
| Description | Eulor5B (Euteleostomi-conserved low frequency repeat 5B) |
| Comment | Putative DNA transposon (assignment unclear). Present in ~50 copies in mammals and chicken, also detected in Xenopus. The sequence is an (imperfect) hairpin. Other members of the Eulor5/6 group all have a non- palindromic region downstream, but this his not been detected in Eulor5B yet. |
| Sequence |
TATTTAAGCAATAATCCCCGAGAAATCGGTCGTTACCAGCAGTTAACGACCGGTTTGTTAGTTAACGGCCCGAGGCGAAGCCGAGGGCGGTTAACGCTCTAACAAACCGGTCGTTAACTGCGGTAACGACCGATTTCGAGGGGATTATTGCTATTATAAACCATANTCAACGGTTTATAACAGCAATAATGT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor5B | MYBL1 | 110 | 121 | + | 11.72 | GTCGTTAACTGC |
| Eulor5B | MYBL1 | 39 | 50 | - | 11.72 | GTCGTTAACTGC |
| Eulor5B | MYB3R3 | 63 | 70 | - | 11.70 | GGCCGTTA |
| Eulor5B | btd | 83 | 91 | + | 11.58 | GAGGGCGGT |
| Eulor5B | MYB65 | 86 | 93 | - | 11.56 | TAACCGCC |
| Eulor5B | PITX2 | 11 | 18 | + | 11.54 | ATAATCCC |
| Eulor5B | PITX2 | 141 | 148 | - | 11.54 | ATAATCCC |
| Eulor5B | Hoxa11 | 110 | 118 | + | 11.53 | GTCGTTAAC |
| Eulor5B | Hoxa11 | 42 | 50 | - | 11.53 | GTCGTTAAC |
| Eulor5B | Zm00001d005692 | 184 | 191 | + | 11.53 | CAATAATG |
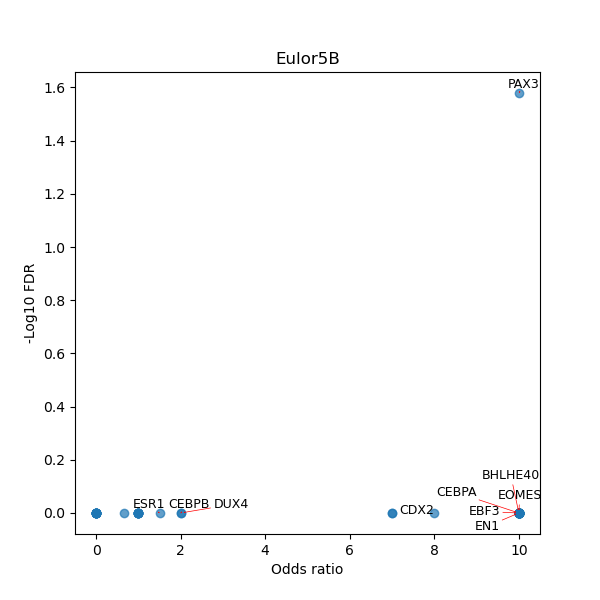
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.