Eulor5A
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000130 |
|---|---|
| TE superfamily | Crypton-A |
| TE class | DNA |
| Species | Tetrapoda |
| Length | 316 |
| Kimura value | 28.98 |
| Tau index | 0.0000 |
| Description | Eulor5A (Euteleostomi-conserved low frequency repeat 5A) |
| Comment | Putative DNA transposon (assignment unclear). Position (roughly) 1-160 is an imperfect hairpin. The hairpin of Eulor5A/B and Eulor6A-E are at the same position and up to 70% similar. The termini are also similar. |
| Sequence |
CTTAATTAAGCAATAACGATCGAGGCGCAGGGCATTTCCTGGGGATTAATGACCGGCTGGGAGGAGTTGATGGCCCGAGGCANAGCCGAGGGCCATTAACCCCAGCCGGTCATTAATCCCCAGGAAATGCCCTGCGCCGAGGTCGTTATTGCTATTATAAGCTGAAAACGNAGAAACGAACAGGCGTATGGATTTTTTTTATGGGTGATGCAGTTTCAATTGGTATGTACAGGGCATTTCCGGAGAATTAATGCCCTGTACATTAGCCAATCAGATTGCTCGAATCATCTCTCAACATTCCATTCGGCTTATAATT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor5A | MEF2D | 151 | 162 | - | 6.38 | GCTTATAATAGC |
| Eulor5A | HSF1 | 277 | 289 | + | 6.07 | TGCTCGAATCATC |
| Eulor5A | RLM1 | 150 | 163 | + | 5.79 | TGCTATTATAAGCT |
| Eulor5A | ZSCAN31 | 245 | 262 | + | 1.60 | GAATTAATGCCCTGTACA |
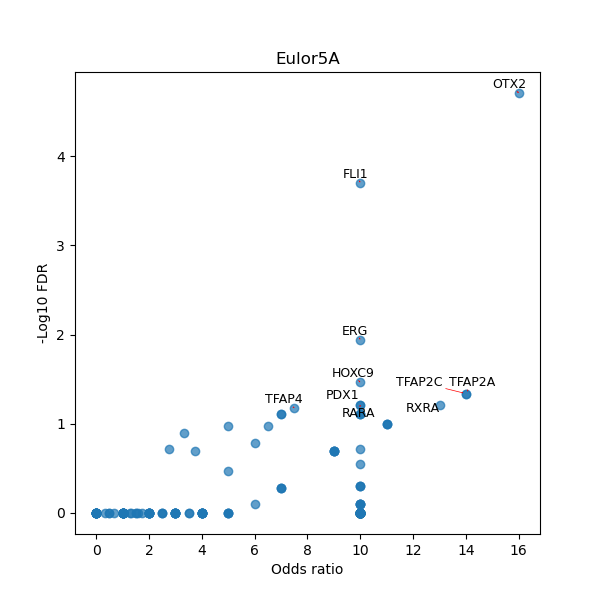
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.