Eulor3
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000128 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 304 |
| Kimura value | 27.98 |
| Tau index | 0.0000 |
| Description | Eulor3 (Euteleostomi-conserved low frequency repeat 3) |
| Comment | Putative DNA transposon (assignment unclear). Like Eulor1 and Eulor2A/B it has a hairpin-like structure followed by a tail. Eulor3 consensus was derived from human copies. Termini are weak and probably incomplete. Near perfect (>90% id) hairpin overall, which contains a number of 41 bp and 82 bp imperfect internal palindromes. Appears in eutherians, platypus, chicken (40-50 copies each), lizard, not in Xenopus. |
| Sequence |
GAACATCTTGGCACTACTATACCTTCTGTCCTGTAATTAAACCGAATCTGCTCCTGGACCCGTTCTTCCCTGGACGGCGCCATCTTTTGAGTAATACATCATCAGAGCGCCTCGCTCTGATGATGTATTACNCTAATACATCATCAGAGCGCCGTCGCTCTGATGATGTATTAGTGTAATACATCATCAGAGCGAGGCNCACTGATGATGTATTACTCAAAAGATGGCGCCGTCCGAGGAAGAAAGGGTCCAGGAGCAGATTCANTNTAATTACAGNATAAAAGNCAACCTAGNGCCAGGAGGC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor3 | ZFP42 | 220 | 232 | + | 13.51 | AAAGATGGCGCCG |
| Eulor3 | ZFP42 | 75 | 87 | - | 13.51 | AAAGATGGCGCCG |
| Eulor3 | NFIC | 7 | 13 | + | 13.37 | CTTGGCA |
| Eulor3 | OsRR22 | 41 | 49 | - | 13.28 | AGATTCGGT |
| Eulor3 | Zm00001d024324 | 150 | 158 | + | 13.14 | CGCCGTCGC |
| Eulor3 | SMZ | 232 | 239 | - | 13.13 | CCTCGGAC |
| Eulor3 | pal-1 | 33 | 40 | + | 13.09 | GTAATTAA |
| Eulor3 | ARR14 | 42 | 49 | - | 12.93 | AGATTCGG |
| Eulor3 | RIM101 | 7 | 13 | - | 12.82 | TGCCAAG |
| Eulor3 | ZHD5 | 33 | 40 | + | 12.67 | GTAATTAA |
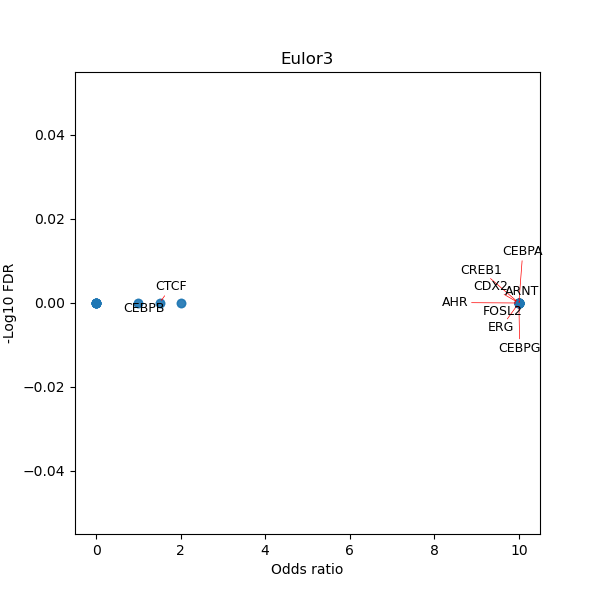
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.