Eulor2C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000127 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 168 |
| Kimura value | 26.12 |
| Tau index | 0.0000 |
| Description | Eulor2C (Euteleostomi-conserved low frequency repeat 2C) |
| Comment | Putative DNA transposon (assignment unclear). Distantly related to Eulor2A and 2B. It has a conserved secondary structure. Matches part of the hairpin of Eulor2A & B, but < 75% similar. |
| Sequence |
TANTTAAGGGATAATGTTCACGGCGGAGGAGTATACGAAGCAATAAACGGCTTTTGCGGGTGATTAAACGCCGAAGTGAAGCTGAGGCGTTTGATNAACCGCAAAAGCCGTTTATTGCGAGTATACTCCAATGCCACGAACATTATTCCTATTACACGACAAGANAGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor2C | ARF16 | 155 | 164 | + | 13.52 | CACGACAAGA |
| Eulor2C | ERF5 | 18 | 32 | - | 13.35 | ACTCCTCCGCCGTGA |
| Eulor2C | CDX4 | 110 | 118 | - | 13.34 | GCAATAAAC |
| Eulor2C | CDX4 | 40 | 48 | + | 13.34 | GCAATAAAC |
| Eulor2C | ERF087 | 19 | 28 | + | 13.27 | CACGGCGGAG |
| Eulor2C | GAF1 | 150 | 164 | + | 13.06 | TATTACACGACAAGA |
| Eulor2C | ERF091 | 20 | 32 | + | 13.06 | ACGGCGGAGGAGT |
| Eulor2C | MATALPHA2 | 151 | 158 | - | 12.89 | CGTGTAAT |
| Eulor2C | ERF095 | 20 | 32 | + | 12.80 | ACGGCGGAGGAGT |
| Eulor2C | Cdx | 111 | 118 | + | 12.76 | TTTATTGC |
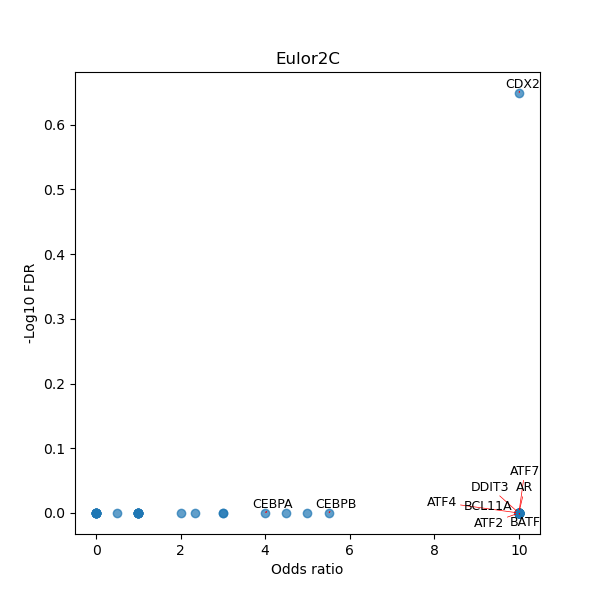
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.