Eulor2B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000126 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 273 |
| Kimura value | 27.92 |
| Tau index | 0.0000 |
| Description | Eulor2B (Euteleostomi-conserved low frequency repeat 2B) |
| Comment | Putative DNA transposon (assignment unclear). Eulor2B has a 34bp internal deletion relative to Eulor2A. Hairpin (roughly) pos 1-157. |
| Sequence |
TAATTAAGAGATAATGTCAATGGAATAGAACGTTGTCACAGGATAATGGTCTCCCGCTGCTAGATAAATGCCGAGGCGNAAGCCGAGACGTTTATTTTCAAAGCAGGAGACATTGATCCTGTGACAACGTTCTATTACAATGACTTTATTTCTATTATACCAAATGATTGATGTAGATTTAATCATTTTGTCTGATGGATGTTGGTGCAGTAGAGTGACAGNTGCTCGCTGTACCATCGTTGANTTGCTGCGTTCGGATTGGCTTAGAAAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor2B | ZSCAN16 | 115 | 132 | - | -20.91 | GAACGTTGTCACAGGATC |
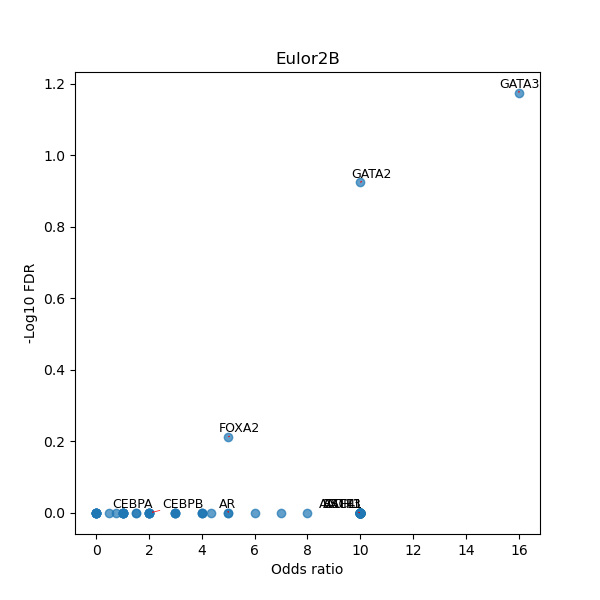
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.