Eulor2A
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000125 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 306 |
| Kimura value | 27.05 |
| Tau index | 0.0000 |
| Description | Eulor2A (Euteleostomi-conserved low frequency repeat 2A) |
| Comment | Putative DNA transposon (assignment unclear). Present in chicken (~200 copies) and mammals (~150 copies). Split into two subfamilies (Eulor2A and 2B), differing mainly by a 31bp indel. Like Eulor1, Eulor2 subfamilies also have a characteristic hairpin-tail structure. Hairpin (roughly) pos 8-188. |
| Sequence |
TAATTAAGAGATAATGTCAATGGAATAGAACGTTGTCACGGGATAATGGTCTCCCGCTGCTAGATAAATGCCGAGGCGAAGCCGAGGCATTTATCGAAAATAAACGTCGAGGCGAAGCCGAGACGTTTATTTTCAAAGCGGGAGACATTGATCCTGTGACAACGTTCTATTACAATGACTTTATTTCTGTTATACCAAATGATTGATGTAGATTTAATCACTTTGTCTGATGGATGTTGGTGCAGCGGAATGACAGTCGCTCGCCGTACCGTTNTTAANCNGCTGCGTTCTGATCGGCTTAGGGGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor2A | egl-38 | 32 | 42 | - | 7.85 | CCCGTGACAAC |
| Eulor2A | IRF7 | 95 | 107 | + | 7.84 | CGAAAATAAACGT |
| Eulor2A | USF2 | 35 | 44 | + | 7.04 | GTCACGGGAT |
| Eulor2A | ZIC1 | 48 | 61 | + | 6.02 | GGTCTCCCGCTGCT |
| Eulor2A | ERF::SREBF2 | 29 | 44 | + | 5.56 | AACGTTGTCACGGGAT |
| Eulor2A | RAX3 | 232 | 243 | + | 4.56 | GGATGTTGGTGC |
| Eulor2A | ZSCAN16 | 150 | 167 | - | -20.91 | GAACGTTGTCACAGGATC |
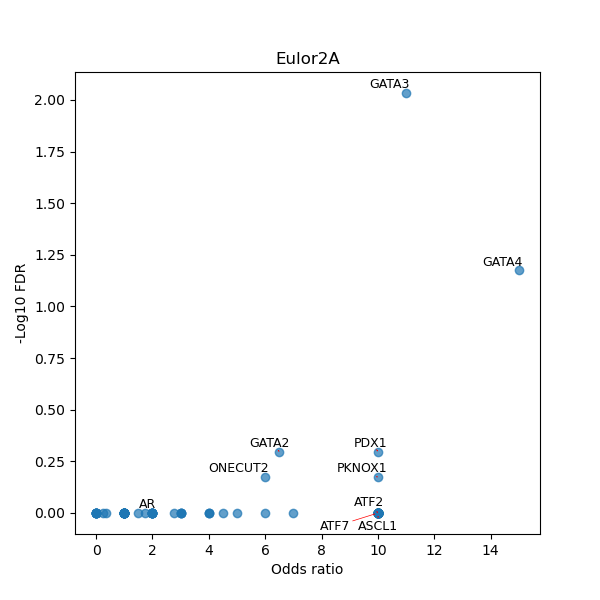
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.