Eulor12
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000124 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 184 |
| Kimura value | 26.22 |
| Tau index | 0.0000 |
| Description | Eulor12 (Euteleostomi-conserved low frequency repeat 12) |
| Comment | Putative DNA transposon (assignment unclear). Identified in both mammals and birds. Reconstructed from chicken DNA. ~100 copies/genome. Palindromic. Appears to be broken up in some subgroups. |
| Sequence |
CATTGNATAAAAAATAANAAATAGCCAACTGTGAATNACGAGGCTGTAATTCCATCTCGGGGTTCCGGTGACGTTAATAAACCGCTCGAGCTTCGCTCTCGTGGTTTACGACGTCACCAGAACCCCTCGATGGAATTACAGCCTTGTAATTCGCAGTTGGCTATNCGTTATTCTGTATGCAATG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor12 | CREB3L4 | 108 | 116 | + | 11.48 | ACGACGTCA |
| Eulor12 | Creb1 | 108 | 117 | + | 11.48 | ACGACGTCAC |
| Eulor12 | Creb1 | 108 | 117 | - | 11.48 | GTGACGTCGT |
| Eulor12 | Lg3 | 110 | 116 | - | 11.44 | TGACGTC |
| Eulor12 | FOSB::JUNB | 108 | 117 | - | 11.38 | GTGACGTCGT |
| Eulor12 | FOS::JUN | 108 | 117 | + | 11.37 | ACGACGTCAC |
| Eulor12 | CREM | 108 | 117 | + | 11.32 | ACGACGTCAC |
| Eulor12 | CREM | 108 | 117 | - | 11.32 | GTGACGTCGT |
| Eulor12 | atf-7 | 68 | 76 | + | 11.31 | GTGACGTTA |
| Eulor12 | FOSB::JUN | 108 | 118 | - | 11.27 | GGTGACGTCGT |
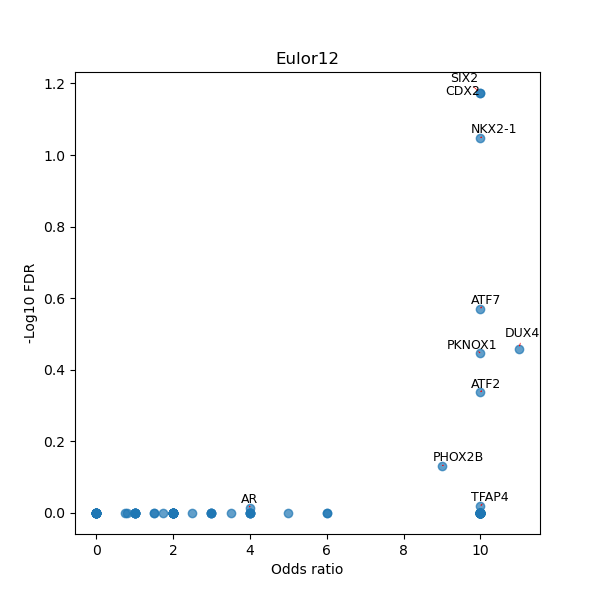
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.