Eulor12
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000124 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 184 |
| Kimura value | 26.22 |
| Tau index | 0.0000 |
| Description | Eulor12 (Euteleostomi-conserved low frequency repeat 12) |
| Comment | Putative DNA transposon (assignment unclear). Identified in both mammals and birds. Reconstructed from chicken DNA. ~100 copies/genome. Palindromic. Appears to be broken up in some subgroups. |
| Sequence |
CATTGNATAAAAAATAANAAATAGCCAACTGTGAATNACGAGGCTGTAATTCCATCTCGGGGTTCCGGTGACGTTAATAAACCGCTCGAGCTTCGCTCTCGTGGTTTACGACGTCACCAGAACCCCTCGATGGAATTACAGCCTTGTAATTCGCAGTTGGCTATNCGTTATTCTGTATGCAATG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor12 | TGA4 | 109 | 119 | + | 15.38 | CGACGTCACCA |
| Eulor12 | Atf-2 | 107 | 117 | + | 14.23 | TACGACGTCAC |
| Eulor12 | TGA1 | 111 | 118 | - | 13.47 | GGTGACGT |
| Eulor12 | TGA1 | 67 | 74 | + | 13.47 | GGTGACGT |
| Eulor12 | HOXC9 | 104 | 112 | - | 13.39 | GTCGTAAAC |
| Eulor12 | TGA10 | 109 | 118 | - | 13.24 | GGTGACGTCG |
| Eulor12 | HOXB9 | 104 | 112 | - | 13.24 | GTCGTAAAC |
| Eulor12 | TGA9 | 108 | 118 | - | 13.08 | GGTGACGTCGT |
| Eulor12 | aop | 63 | 69 | - | 12.82 | ACCGGAA |
| Eulor12 | JUN::JUNB | 108 | 118 | - | 12.78 | GGTGACGTCGT |
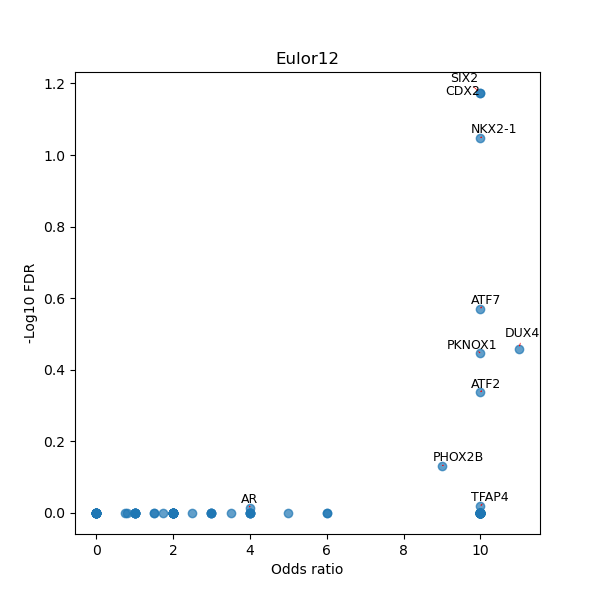
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.