Eulor12
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000124 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 184 |
| Kimura value | 26.22 |
| Tau index | 0.0000 |
| Description | Eulor12 (Euteleostomi-conserved low frequency repeat 12) |
| Comment | Putative DNA transposon (assignment unclear). Identified in both mammals and birds. Reconstructed from chicken DNA. ~100 copies/genome. Palindromic. Appears to be broken up in some subgroups. |
| Sequence |
CATTGNATAAAAAATAANAAATAGCCAACTGTGAATNACGAGGCTGTAATTCCATCTCGGGGTTCCGGTGACGTTAATAAACCGCTCGAGCTTCGCTCTCGTGGTTTACGACGTCACCAGAACCCCTCGATGGAATTACAGCCTTGTAATTCGCAGTTGGCTATNCGTTATTCTGTATGCAATG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor12 | ERT1 | 62 | 68 | - | 12.73 | CCGGAAC |
| Eulor12 | Hoxa11 | 104 | 112 | - | 12.71 | GTCGTAAAC |
| Eulor12 | CREB3L4 | 109 | 117 | - | 12.58 | GTGACGTCG |
| Eulor12 | PK09021.1 | 109 | 116 | - | 12.57 | TGACGTCG |
| Eulor12 | Atf6 | 107 | 119 | - | 12.52 | TGGTGACGTCGTA |
| Eulor12 | atf-7 | 109 | 117 | - | 12.34 | GTGACGTCG |
| Eulor12 | HOXD11 | 104 | 112 | - | 12.33 | GTCGTAAAC |
| Eulor12 | HOXD12 | 104 | 113 | - | 12.30 | CGTCGTAAAC |
| Eulor12 | HOXD10 | 104 | 113 | - | 12.29 | CGTCGTAAAC |
| Eulor12 | FOS | 106 | 118 | - | 11.86 | GGTGACGTCGTAA |
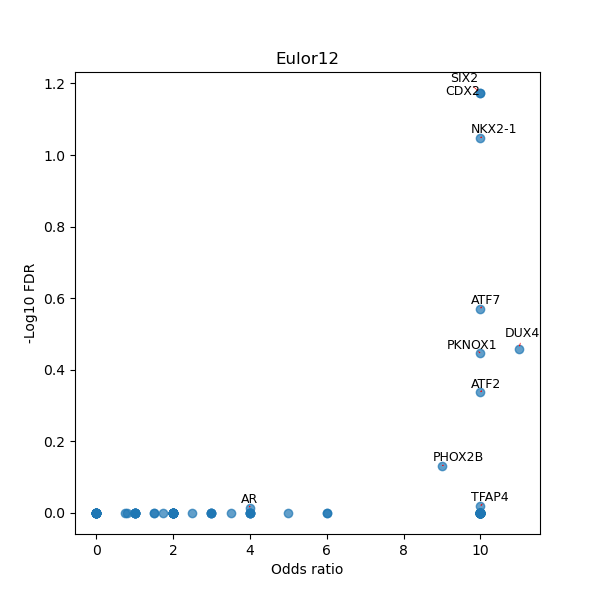
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.