Eulor10
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000122 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 317 |
| Kimura value | 25.37 |
| Tau index | 0.0000 |
| Description | Eulor10 (Euteleostomi-conserved low frequency repeat 10) |
| Comment | Putative DNA transposon (assignment unclear). Termini unclear. Eulor10 is present in mammals and birds (~40 copies), and alligator (~100 copies). Follows characteristic hairpin-tail structure, with at least two internal palindromes. |
| Sequence |
TGAAACGCGGGNCGGATTCATCAAAGGAGAGCTGGNANTTACCGACTCCGNATATTTTGCTTNAATTTTTTTTNTCTGATTCATGAANCGCTTTTCGAAATTCGAAAAGCGGTTCATGAATCGNTCGGGAGCCGGCAAAAATTAATAGTAATGAGCTCATTTCCATAGAAATGGGCTCATTACCATGCCGGCTGCCGAAAATTTACGAGGCCGGNTTCTTGCCGGCAAACGGGGTCCTGCAGGCAGTGTCCTTCCGCCTGCCTGCAGGAAATAATCCCNTNTTNAAAAATTAANCAAAAACAAATTTTAGTTTATTT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor10 | ERF6 | 221 | 227 | - | 13.72 | TGCCGGC |
| Eulor10 | PK24580.1 | 131 | 137 | + | 13.69 | GCCGGCA |
| Eulor10 | PK24580.1 | 186 | 192 | - | 13.69 | GCCGGCA |
| Eulor10 | PK24580.1 | 220 | 226 | - | 13.69 | GCCGGCA |
| Eulor10 | PK24580.1 | 221 | 227 | + | 13.69 | GCCGGCA |
| Eulor10 | ZNF384 | 66 | 73 | - | 13.57 | AAAAAAAA |
| Eulor10 | Hnf1A | 18 | 27 | - | 13.29 | CCTTTGATGA |
| Eulor10 | POU6F2 | 174 | 182 | + | 13.20 | GGCTCATTA |
| Eulor10 | nub | 157 | 167 | - | 12.97 | TATGGAAATGA |
| Eulor10 | JUND | 150 | 160 | + | 12.68 | AATGAGCTCAT |
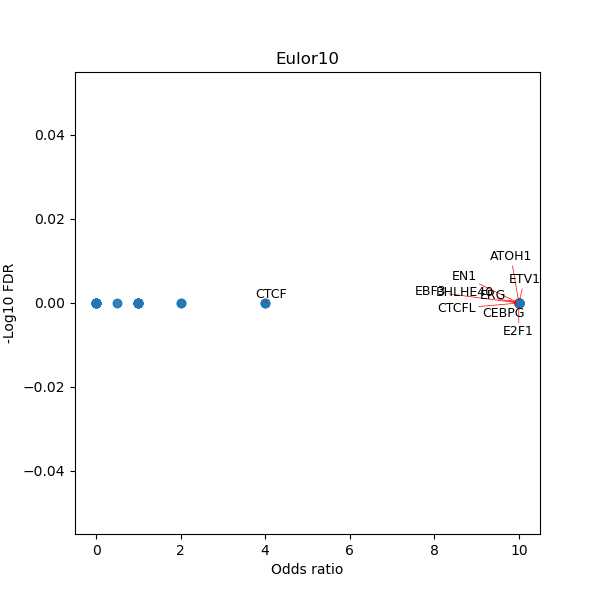
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.